Gene Page: PPFIA2
Summary ?
| GeneID | 8499 |
| Symbol | PPFIA2 |
| Synonyms | - |
| Description | PTPRF interacting protein alpha 2 |
| Reference | MIM:603143|HGNC:HGNC:9246|Ensembl:ENSG00000139220|HPRD:04392|Vega:OTTHUMG00000170181 |
| Gene type | protein-coding |
| Map location | 12q21.31 |
| Pascal p-value | 0.365 |
| Fetal beta | 0.168 |
| eGene | Caudate basal ganglia |
| Support | PROTEIN CLUSTERING G2Cdb.humanPSD G2Cdb.humanPSP CompositeSet Potential synaptic genes |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CV:GWAScat | Genome-wide Association Studies | This data set includes 560 SNPs associated with schizophrenia. A total of 486 genes were mapped to these SNPs within 50kb. | |
| CV:GWASdb | Genome-wide Association Studies | GWASdb records for schizophrenia | |
| CV:PGCnp | Genome-wide Association Study | GWAS | |
| GO_Annotation | Mapping neuro-related keywords to Gene Ontology annotations | Hits with neuro-related keywords: 1 |
Section I. Genetics and epigenetics annotation
Section II. Transcriptome annotation
General gene expression (GTEx)
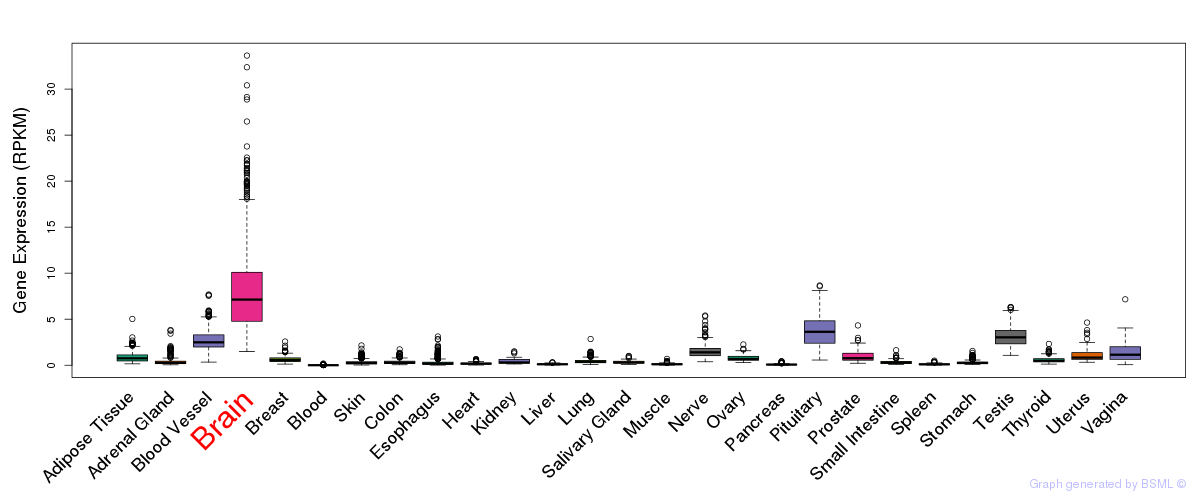
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
Top co-expressed genes in brain regions
| Top 10 positively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| MAPRE3 | 0.87 | 0.88 |
| ATP6V0A1 | 0.87 | 0.88 |
| WARS | 0.86 | 0.85 |
| BECN1 | 0.86 | 0.84 |
| AARS | 0.86 | 0.88 |
| PDHX | 0.86 | 0.83 |
| DDX24 | 0.86 | 0.81 |
| UQCC | 0.85 | 0.85 |
| PCMT1 | 0.85 | 0.87 |
| NDEL1 | 0.85 | 0.85 |
| Top 10 negatively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| AF347015.21 | -0.55 | -0.30 |
| AF347015.18 | -0.54 | -0.30 |
| AP002478.3 | -0.51 | -0.44 |
| AF347015.2 | -0.51 | -0.28 |
| RAB34 | -0.49 | -0.53 |
| AF347015.8 | -0.47 | -0.26 |
| AF347015.31 | -0.47 | -0.30 |
| IL32 | -0.47 | -0.29 |
| MT-CO2 | -0.46 | -0.28 |
| AC100783.1 | -0.46 | -0.34 |
Section III. Gene Ontology annotation
| Molecular function | GO term | Evidence | Neuro keywords | PubMed ID |
|---|---|---|---|---|
| GO:0005515 | protein binding | IPI | 12923177 | |
| Biological process | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0007160 | cell-matrix adhesion | TAS | 9624153 | |
| Cellular component | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0019717 | synaptosome | IEA | Synap, Brain (GO term level: 7) | - |
| GO:0005737 | cytoplasm | TAS | 9624153 | |
| GO:0009986 | cell surface | IEA | - |
Section IV. Protein-protein interaction annotation
| Interactors | Aliases B | Official full name B | Experimental | Source | PubMed ID |
|---|---|---|---|---|---|
| ERC2 | CAST | CAST1 | ELKSL | KIAA0378 | MGC133063 | MGC133064 | SPBC110 | Spc110 | ELKS/RAB6-interacting/CAST family member 2 | - | HPRD,BioGRID | 12923177 |
| ERC2 | CAST | CAST1 | ELKSL | KIAA0378 | MGC133063 | MGC133064 | SPBC110 | Spc110 | ELKS/RAB6-interacting/CAST family member 2 | Liprin-alpha2 interacts with ERC2. | BIND | 12923177 |
| GIT1 | - | G protein-coupled receptor kinase interacting ArfGAP 1 | Reconstituted Complex Two-hybrid | BioGRID | 12923177 |
| GIT1 | - | G protein-coupled receptor kinase interacting ArfGAP 1 | - | HPRD | 12629171 |
| GIT1 | - | G protein-coupled receptor kinase interacting ArfGAP 1 | Liprin-alpha2 interacts with GIT1. This interaction was modeled on a demonstrated interaction between human liprin-alpha2 and rat GIT1. | BIND | 12923177 |
| PPFIA1 | FLJ41337 | FLJ42630 | FLJ43474 | LIP.1 | LIP1 | LIPRIN | MGC26800 | protein tyrosine phosphatase, receptor type, f polypeptide (PTPRF), interacting protein (liprin), alpha 1 | - | HPRD,BioGRID | 9624153 |
| PPFIA2 | FLJ41378 | MGC132572 | protein tyrosine phosphatase, receptor type, f polypeptide (PTPRF), interacting protein (liprin), alpha 2 | - | HPRD,BioGRID | 9624153 |
| PPFIA3 | KIAA0654 | LPNA3 | MGC126567 | MGC126569 | protein tyrosine phosphatase, receptor type, f polypeptide (PTPRF), interacting protein (liprin), alpha 3 | - | HPRD,BioGRID | 9624153 |
| PPFIBP1 | L2 | hSGT2 | hSgt2p | PTPRF interacting protein, binding protein 1 (liprin beta 1) | - | HPRD,BioGRID | 9624153 |
| PPFIBP2 | Cclp1 | DKFZp781K06126 | MGC42541 | PTPRF interacting protein, binding protein 2 (liprin beta 2) | - | HPRD,BioGRID | 9624153 |
| PPP2CA | PP2Ac | PP2CA | RP-C | protein phosphatase 2 (formerly 2A), catalytic subunit, alpha isoform | Affinity Capture-MS | BioGRID | 18782753 |
| PPP2R1A | MGC786 | PR65A | protein phosphatase 2 (formerly 2A), regulatory subunit A, alpha isoform | Affinity Capture-MS | BioGRID | 18782753 |
| PTPRD | HPTP | HPTP-DELTA | HPTPD | MGC119750 | MGC119751 | MGC119752 | MGC119753 | PTPD | R-PTP-DELTA | protein tyrosine phosphatase, receptor type, D | - | HPRD,BioGRID | 9624153 |
| PTPRF | FLJ43335 | FLJ45062 | FLJ45567 | LAR | protein tyrosine phosphatase, receptor type, F | - | HPRD,BioGRID | 9624153 |
| PTPRS | PTPSIGMA | protein tyrosine phosphatase, receptor type, S | - | HPRD,BioGRID | 9624153 |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| ZHOU INFLAMMATORY RESPONSE LPS UP | 431 | 237 | All SZGR 2.0 genes in this pathway |
| BROWNE HCMV INFECTION 30MIN DN | 150 | 99 | All SZGR 2.0 genes in this pathway |
| VERHAAK GLIOBLASTOMA NEURAL | 129 | 85 | All SZGR 2.0 genes in this pathway |
Section VI. microRNA annotation
| miRNA family | Target position | miRNA ID | miRNA seq | ||
|---|---|---|---|---|---|
| UTR start | UTR end | Match method | |||
| miR-130/301 | 65 | 71 | 1A | hsa-miR-130abrain | CAGUGCAAUGUUAAAAGGGCAU |
| hsa-miR-301 | CAGUGCAAUAGUAUUGUCAAAGC | ||||
| hsa-miR-130bbrain | CAGUGCAAUGAUGAAAGGGCAU | ||||
| hsa-miR-454-3p | UAGUGCAAUAUUGCUUAUAGGGUUU | ||||
| miR-148/152 | 21 | 27 | 1A | hsa-miR-148a | UCAGUGCACUACAGAACUUUGU |
| hsa-miR-152brain | UCAGUGCAUGACAGAACUUGGG | ||||
| hsa-miR-148b | UCAGUGCAUCACAGAACUUUGU | ||||
- SZ: miRNAs which differentially expressed in brain cortex of schizophrenia patients comparing with control samples using microarray. Click here to see the list of SZ related miRNAs.
- Brain: miRNAs which are expressed in brain based on miRNA microarray expression studies. Click here to see the list of brain related miRNAs.