Gene Page: CAV3
Summary ?
| GeneID | 859 |
| Symbol | CAV3 |
| Synonyms | LGMD1C|LQT9|VIP-21|VIP21 |
| Description | caveolin 3 |
| Reference | MIM:601253|HGNC:HGNC:1529|Ensembl:ENSG00000182533|HPRD:03154|Vega:OTTHUMG00000090519 |
| Gene type | protein-coding |
| Map location | 3p25 |
| Pascal p-value | 0.497 |
| Fetal beta | -0.122 |
| DMG | 1 (# studies) |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CV:PGCnp | Genome-wide Association Study | GWAS | |
| DMG:Wockner_2014 | Genome-wide DNA methylation analysis | This dataset includes 4641 differentially methylated probes corresponding to 2929 unique genes between schizophrenia patients (n=24) and controls (n=24). | 1 |
| PMID:cooccur | High-throughput literature-search | Systematic search in PubMed for genes co-occurring with SCZ keywords. A total of 3027 genes were included. | |
| Literature | High-throughput literature-search | Co-occurance with Schizophrenia keywords: schizophrenia,schizophrenias | Click to show details |
Section I. Genetics and epigenetics annotation
 Differentially methylated gene
Differentially methylated gene
| Probe | Chromosome | Position | Nearest gene | P (dis) | Beta (dis) | FDR (dis) | Study |
|---|---|---|---|---|---|---|---|
| cg16448890 | 3 | 8783613 | CAV3 | 3.419E-4 | -0.303 | 0.041 | DMG:Wockner_2014 |
Section II. Transcriptome annotation
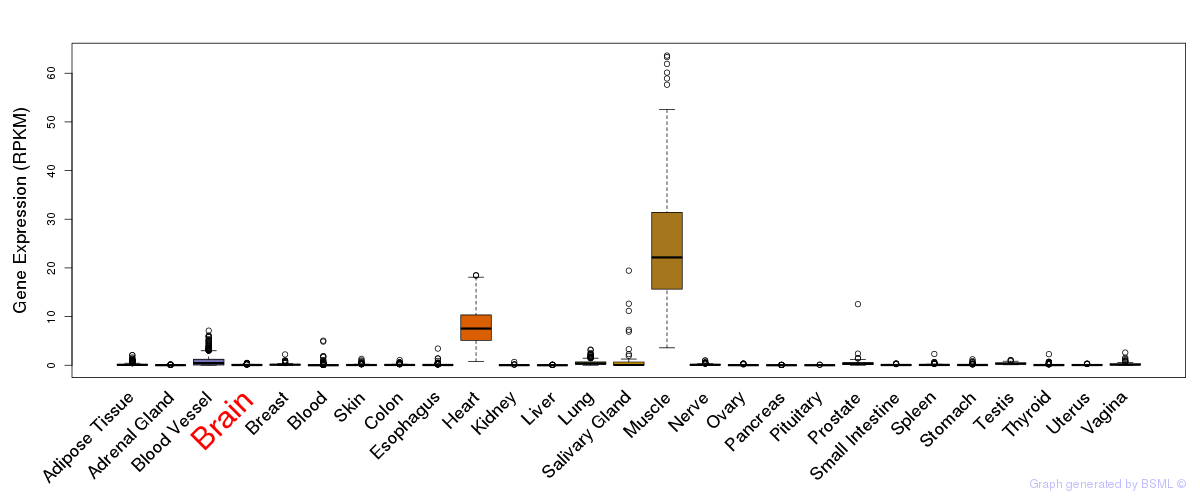
General gene expression (GTEx)
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
Top co-expressed genes in brain regions
| Top 10 positively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| ZNF710 | 0.90 | 0.90 |
| CACNA1H | 0.89 | 0.90 |
| TNRC18B | 0.89 | 0.88 |
| BAHCC1 | 0.88 | 0.90 |
| RFX1 | 0.87 | 0.91 |
| RAI1 | 0.86 | 0.92 |
| ELAVL3 | 0.86 | 0.91 |
| FAM155B | 0.85 | 0.85 |
| ZBED1 | 0.85 | 0.84 |
| NTN3 | 0.85 | 0.77 |
| Top 10 negatively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| AF347015.31 | -0.62 | -0.83 |
| C5orf53 | -0.60 | -0.71 |
| MT-CO2 | -0.60 | -0.81 |
| AF347015.27 | -0.59 | -0.79 |
| S100B | -0.59 | -0.77 |
| COPZ2 | -0.59 | -0.75 |
| AF347015.33 | -0.58 | -0.76 |
| MT-CYB | -0.58 | -0.78 |
| FXYD1 | -0.57 | -0.76 |
| AF347015.8 | -0.57 | -0.79 |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| KEGG FOCAL ADHESION | 201 | 138 | All SZGR 2.0 genes in this pathway |
| ST MYOCYTE AD PATHWAY | 27 | 25 | All SZGR 2.0 genes in this pathway |
| PID PDGFRA PATHWAY | 22 | 18 | All SZGR 2.0 genes in this pathway |
| GRAESSMANN APOPTOSIS BY SERUM DEPRIVATION DN | 234 | 147 | All SZGR 2.0 genes in this pathway |
| MARTINEZ RB1 TARGETS UP | 673 | 430 | All SZGR 2.0 genes in this pathway |
| MARTINEZ TP53 TARGETS UP | 602 | 364 | All SZGR 2.0 genes in this pathway |
| MARTINEZ RB1 AND TP53 TARGETS UP | 601 | 369 | All SZGR 2.0 genes in this pathway |
| LABBE WNT3A TARGETS UP | 112 | 71 | All SZGR 2.0 genes in this pathway |
| SWEET LUNG CANCER KRAS DN | 435 | 289 | All SZGR 2.0 genes in this pathway |
| BROWNE HCMV INFECTION 30MIN UP | 56 | 38 | All SZGR 2.0 genes in this pathway |
| BRUINS UVC RESPONSE VIA TP53 GROUP A | 898 | 516 | All SZGR 2.0 genes in this pathway |
| WANG NFKB TARGETS | 25 | 15 | All SZGR 2.0 genes in this pathway |