Gene Page: TNKS
Summary ?
| GeneID | 8658 |
| Symbol | TNKS |
| Synonyms | ARTD5|PARP-5a|PARP5A|PARPL|TIN1|TINF1|TNKS1|pART5 |
| Description | tankyrase |
| Reference | MIM:603303|HGNC:HGNC:11941|Ensembl:ENSG00000173273|HPRD:04490|Vega:OTTHUMG00000090481 |
| Gene type | protein-coding |
| Map location | 8p23.1 |
| Pascal p-value | 2.426E-5 |
| Fetal beta | 0.981 |
| eGene | Caudate basal ganglia Putamen basal ganglia |
| Support | CompositeSet Darnell FMRP targets |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CV:GWASdb | Genome-wide Association Studies | GWASdb records for schizophrenia | |
| CV:PGCnp | Genome-wide Association Study | GWAS | |
| DNM:Fromer_2014 | Whole Exome Sequencing analysis | This study reported a WES study of 623 schizophrenia trios, reporting DNMs using genomic DNA. |
Section I. Genetics and epigenetics annotation
 DNM table
DNM table
| Gene | Chromosome | Position | Ref | Alt | Transcript | AA change | Mutation type | Sift | CG46 | Trait | Study |
|---|---|---|---|---|---|---|---|---|---|---|---|
| TNKS | chr8 | 9413678 | A | T | NM_003747 | p.77R>W | missense | Schizophrenia | DNM:Fromer_2014 |
 eQTL annotation
eQTL annotation
| SNP ID | Chromosome | Position | eGene | Gene Entrez ID | pvalue | qvalue | TSS distance | eQTL type |
|---|---|---|---|---|---|---|---|---|
| rs13270518 | 8 | 9536713 | TNKS | ENSG00000173273.11 | 1.93467E-6 | 0.05 | 123289 | gtex_brain_putamen_basal |
Section II. Transcriptome annotation
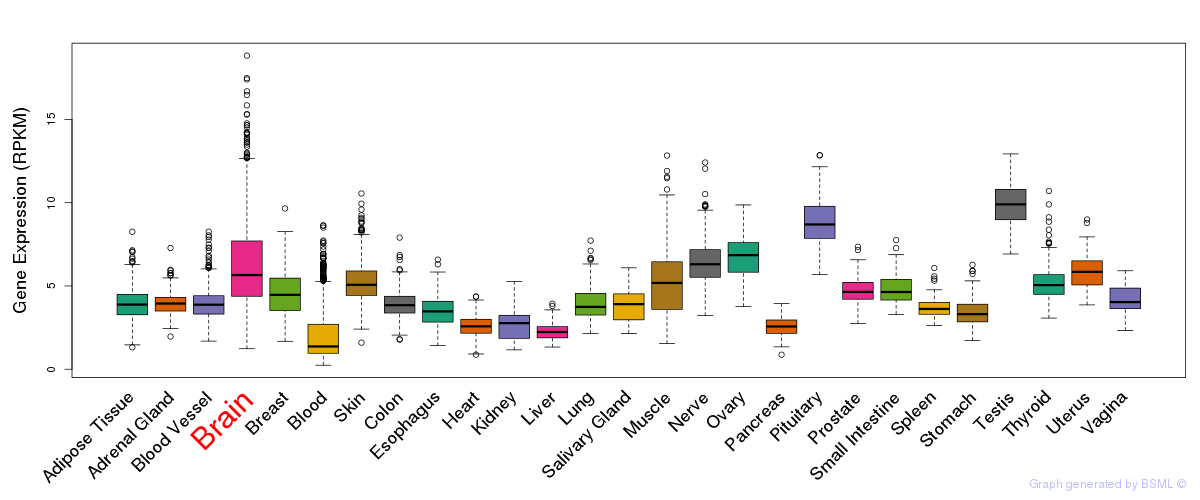
General gene expression (GTEx)
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
Top co-expressed genes in brain regions
| Top 10 positively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| AP3D1 | 0.94 | 0.94 |
| SIN3B | 0.94 | 0.94 |
| UBE2O | 0.94 | 0.94 |
| GGA3 | 0.93 | 0.94 |
| GTF3C1 | 0.93 | 0.94 |
| DEPDC5 | 0.93 | 0.93 |
| HERC2 | 0.93 | 0.93 |
| RUSC2 | 0.93 | 0.94 |
| ZNF142 | 0.93 | 0.95 |
| MGRN1 | 0.93 | 0.94 |
| Top 10 negatively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| AF347015.31 | -0.79 | -0.79 |
| MT-CO2 | -0.78 | -0.79 |
| AF347015.21 | -0.78 | -0.86 |
| HIGD1B | -0.75 | -0.78 |
| FXYD1 | -0.75 | -0.75 |
| C1orf54 | -0.75 | -0.86 |
| AF347015.33 | -0.74 | -0.74 |
| AF347015.8 | -0.73 | -0.76 |
| MT-CYB | -0.73 | -0.74 |
| AF347015.27 | -0.73 | -0.75 |
Section IV. Protein-protein interaction annotation
| Interactors | Aliases B | Official full name B | Experimental | Source | PubMed ID |
|---|---|---|---|---|---|
| FNBP1 | FBP17 | KIAA0554 | MGC126804 | formin binding protein 1 | - | HPRD,BioGRID | 14596906 |
| FNBP1 | FBP17 | KIAA0554 | MGC126804 | formin binding protein 1 | FBP17 interacts with TNKS. | BIND | 14596906 |
| IL1RN | ICIL-1RA | IL-1ra3 | IL1F3 | IL1RA | IRAP | MGC10430 | interleukin 1 receptor antagonist | - | HPRD,BioGRID | 10988299 |11802774 |
| LNPEP | CAP | IRAP | P-LAP | PLAP | leucyl/cystinyl aminopeptidase | - | HPRD,BioGRID | 10988299 |11802774 |
| MCL1 | BCL2L3 | EAT | MCL1L | MCL1S | MGC104264 | MGC1839 | TM | myeloid cell leukemia sequence 1 (BCL2-related) | - | HPRD,BioGRID | 12475993 |
| NUMA1 | NUMA | nuclear mitotic apparatus protein 1 | - | HPRD,BioGRID | 12080061 |
| POT1 | DKFZp586D211 | hPot1 | POT1 protection of telomeres 1 homolog (S. pombe) | - | HPRD,BioGRID | 12768206 |
| TERF1 | FLJ41416 | PIN2 | TRBF1 | TRF | TRF1 | hTRF1-AS | t-TRF1 | telomeric repeat binding factor (NIMA-interacting) 1 | - | HPRD | 9822378 |10988299 |11739745 |11854288 |12080061 |
| TERF1 | FLJ41416 | PIN2 | TRBF1 | TRF | TRF1 | hTRF1-AS | t-TRF1 | telomeric repeat binding factor (NIMA-interacting) 1 | Affinity Capture-Western Biochemical Activity Reconstituted Complex Two-hybrid | BioGRID | 9822378 |11739745 |11854288 |12080061 |
| TNKS | PARP-5a | PARP5A | PARPL | TIN1 | TINF1 | TNKS1 | tankyrase, TRF1-interacting ankyrin-related ADP-ribose polymerase | Biochemical Activity | BioGRID | 9822378 |
| TNKS1BP1 | FLJ45975 | KIAA1741 | TAB182 | tankyrase 1 binding protein 1, 182kDa | - | HPRD,BioGRID | 11854288 |12080061 |
| TNKS2 | PARP-5b | PARP-5c | PARP5B | PARP5C | TANK2 | TNKL | tankyrase, TRF1-interacting ankyrin-related ADP-ribose polymerase 2 | - | HPRD,BioGRID | 11802774 |
| VPS33A | FLJ22395 | FLJ23187 | vacuolar protein sorting 33 homolog A (S. cerevisiae) | - | HPRD | 11382755 |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| BIOCARTA TEL PATHWAY | 18 | 15 | All SZGR 2.0 genes in this pathway |
| PID TELOMERASE PATHWAY | 68 | 48 | All SZGR 2.0 genes in this pathway |
| OSMAN BLADDER CANCER DN | 406 | 230 | All SZGR 2.0 genes in this pathway |
| ENK UV RESPONSE EPIDERMIS UP | 293 | 179 | All SZGR 2.0 genes in this pathway |
| CONCANNON APOPTOSIS BY EPOXOMICIN UP | 239 | 157 | All SZGR 2.0 genes in this pathway |
| DACOSTA UV RESPONSE VIA ERCC3 DN | 855 | 609 | All SZGR 2.0 genes in this pathway |
| AMUNDSON RESPONSE TO ARSENITE | 217 | 143 | All SZGR 2.0 genes in this pathway |
| PUJANA ATM PCC NETWORK | 1442 | 892 | All SZGR 2.0 genes in this pathway |
| ZHAN MULTIPLE MYELOMA CD1 AND CD2 UP | 89 | 51 | All SZGR 2.0 genes in this pathway |
| BROWNE HCMV INFECTION 20HR DN | 101 | 70 | All SZGR 2.0 genes in this pathway |
| LEIN MEDULLA MARKERS | 81 | 48 | All SZGR 2.0 genes in this pathway |
| BONOME OVARIAN CANCER SURVIVAL SUBOPTIMAL DEBULKING | 510 | 309 | All SZGR 2.0 genes in this pathway |
| RUIZ TNC TARGETS UP | 153 | 107 | All SZGR 2.0 genes in this pathway |
| CAIRO HEPATOBLASTOMA UP | 207 | 143 | All SZGR 2.0 genes in this pathway |