Gene Page: FCGBP
Summary ?
| GeneID | 8857 |
| Symbol | FCGBP |
| Synonyms | FC(GAMMA)BP |
| Description | Fc fragment of IgG binding protein |
| Reference | HGNC:HGNC:13572|Ensembl:ENSG00000275395|HPRD:10956|Vega:OTTHUMG00000182580 |
| Gene type | protein-coding |
| Map location | 19q13.1 |
| Pascal p-value | 0.166 |
| Sherlock p-value | 0.094 |
| Fetal beta | 0.669 |
| DMG | 1 (# studies) |
| eGene | Caudate basal ganglia Cortex Hippocampus Nucleus accumbens basal ganglia Myers' cis & trans |
| Support | CompositeSet |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CV:PGCnp | Genome-wide Association Study | GWAS | |
| DMG:Wockner_2014 | Genome-wide DNA methylation analysis | This dataset includes 4641 differentially methylated probes corresponding to 2929 unique genes between schizophrenia patients (n=24) and controls (n=24). | 2 |
| DNM:Xu_2012 | Whole Exome Sequencing analysis | De novo mutations of 4 genes were identified by exome sequencing of 795 samples in this study |
Section I. Genetics and epigenetics annotation
 DNM table
DNM table
| Gene | Chromosome | Position | Ref | Alt | Transcript | AA change | Mutation type | Sift | CG46 | Trait | Study |
|---|---|---|---|---|---|---|---|---|---|---|---|
| FCGBP | chr19 | 40430427 | C | A | NM_003890 | p.506D>Y | missense | Schizophrenia | DNM:Xu_2012 |
 Differentially methylated gene
Differentially methylated gene
| Probe | Chromosome | Position | Nearest gene | P (dis) | Beta (dis) | FDR (dis) | Study |
|---|---|---|---|---|---|---|---|
| cg18051762 | 19 | 40388677 | FCGBP | 1.517E-4 | -0.366 | 0.032 | DMG:Wockner_2014 |
| cg24864275 | 19 | 40421453 | FCGBP | 1.606E-4 | 0.233 | 0.032 | DMG:Wockner_2014 |
 eQTL annotation
eQTL annotation
| SNP ID | Chromosome | Position | eGene | Gene Entrez ID | pvalue | qvalue | TSS distance | eQTL type |
|---|---|---|---|---|---|---|---|---|
| rs473411 | chr1 | 110846615 | FCGBP | 8857 | 0.19 | trans | ||
| rs17004962 | chr4 | 81563338 | FCGBP | 8857 | 0.08 | trans | ||
| rs16873760 | chr6 | 45729388 | FCGBP | 8857 | 0.08 | trans | ||
| rs16873861 | chr6 | 45752854 | FCGBP | 8857 | 0.03 | trans |
Section II. Transcriptome annotation
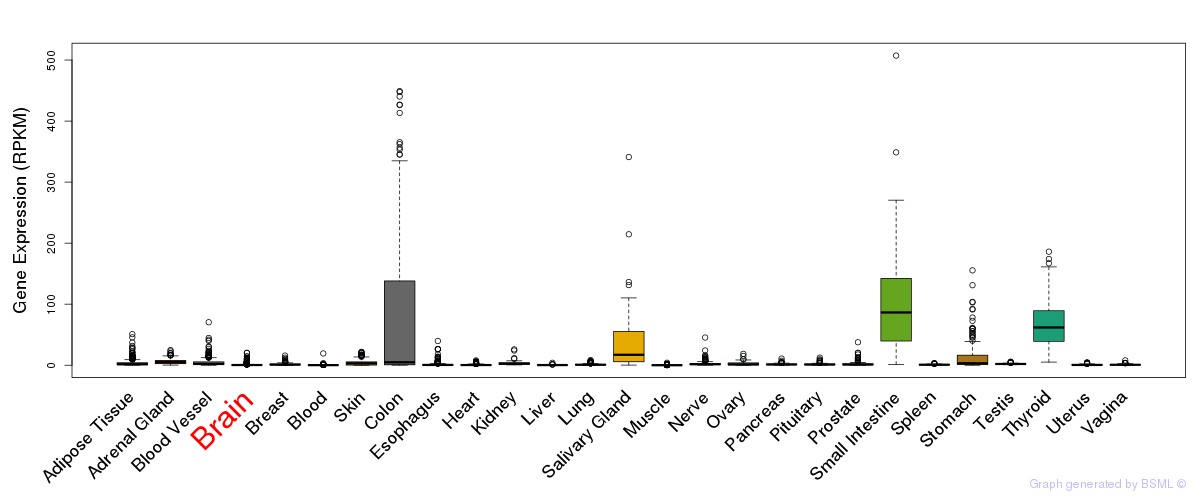
General gene expression (GTEx)
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
Top co-expressed genes in brain regions
| Top 10 positively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| ACVR1B | 0.89 | 0.92 |
| OSBPL10 | 0.89 | 0.89 |
| GPD1L | 0.89 | 0.88 |
| USP6NL | 0.89 | 0.89 |
| SPTAN1 | 0.88 | 0.88 |
| LMO7 | 0.88 | 0.86 |
| CTNND2 | 0.87 | 0.83 |
| HERC3 | 0.87 | 0.89 |
| C5orf25 | 0.86 | 0.89 |
| SEC31A | 0.86 | 0.90 |
| Top 10 negatively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| SERPINB6 | -0.73 | -0.78 |
| HSD17B14 | -0.72 | -0.83 |
| MT-CO2 | -0.72 | -0.83 |
| AF347015.31 | -0.72 | -0.82 |
| FXYD1 | -0.70 | -0.83 |
| HIGD1B | -0.70 | -0.84 |
| AF347015.33 | -0.69 | -0.78 |
| MT-CYB | -0.69 | -0.79 |
| IFI27 | -0.68 | -0.81 |
| AF347015.8 | -0.68 | -0.81 |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| LIU PROSTATE CANCER DN | 481 | 290 | All SZGR 2.0 genes in this pathway |
| DEURIG T CELL PROLYMPHOCYTIC LEUKEMIA UP | 368 | 234 | All SZGR 2.0 genes in this pathway |
| JAEGER METASTASIS DN | 258 | 141 | All SZGR 2.0 genes in this pathway |
| TAKEDA TARGETS OF NUP98 HOXA9 FUSION 16D DN | 143 | 83 | All SZGR 2.0 genes in this pathway |
| ENK UV RESPONSE EPIDERMIS DN | 508 | 354 | All SZGR 2.0 genes in this pathway |
| DELYS THYROID CANCER DN | 232 | 154 | All SZGR 2.0 genes in this pathway |
| WANG BARRETTS ESOPHAGUS UP | 51 | 27 | All SZGR 2.0 genes in this pathway |
| WATTEL AUTONOMOUS THYROID ADENOMA UP | 73 | 47 | All SZGR 2.0 genes in this pathway |
| KOYAMA SEMA3B TARGETS UP | 292 | 168 | All SZGR 2.0 genes in this pathway |
| SANSOM APC TARGETS DN | 366 | 238 | All SZGR 2.0 genes in this pathway |
| CREIGHTON ENDOCRINE THERAPY RESISTANCE 3 | 720 | 440 | All SZGR 2.0 genes in this pathway |
| KUUSELO PANCREATIC CANCER 19Q13 AMPLIFICATION | 35 | 22 | All SZGR 2.0 genes in this pathway |
| FONTAINE FOLLICULAR THYROID ADENOMA UP | 75 | 43 | All SZGR 2.0 genes in this pathway |
| FONTAINE PAPILLARY THYROID CARCINOMA DN | 80 | 53 | All SZGR 2.0 genes in this pathway |
| HOSHIDA LIVER CANCER SUBCLASS S1 | 237 | 159 | All SZGR 2.0 genes in this pathway |