Gene Page: TOP3B
Summary ?
| GeneID | 8940 |
| Symbol | TOP3B |
| Synonyms | TOP3B1 |
| Description | topoisomerase (DNA) III beta |
| Reference | MIM:603582|HGNC:HGNC:11993|Ensembl:ENSG00000100038|HPRD:04662|Vega:OTTHUMG00000167438 |
| Gene type | protein-coding |
| Map location | 22q11.22 |
| Pascal p-value | 0.015 |
| Sherlock p-value | 0.301 |
| Fetal beta | -0.34 |
| eGene | Anterior cingulate cortex BA24 Caudate basal ganglia Cerebellar Hemisphere Cerebellum Cortex Frontal Cortex BA9 Hippocampus Nucleus accumbens basal ganglia |
| Support | CompositeSet Ascano FMRP targets |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CV:PGCnp | Genome-wide Association Study | GWAS | |
| DNM:Xu_2012 | Whole Exome Sequencing analysis | De novo mutations of 4 genes were identified by exome sequencing of 795 samples in this study | |
| PMID:cooccur | High-throughput literature-search | Systematic search in PubMed for genes co-occurring with SCZ keywords. A total of 3027 genes were included. | |
| GSMA_I | Genome scan meta-analysis | Psr: 0.031 | |
| Literature | High-throughput literature-search | Co-occurance with Schizophrenia keywords: schizophrenia,schizophrenias | Click to show details |
Section I. Genetics and epigenetics annotation
 DNM table
DNM table
| Gene | Chromosome | Position | Ref | Alt | Transcript | AA change | Mutation type | Sift | CG46 | Trait | Study |
|---|---|---|---|---|---|---|---|---|---|---|---|
| TOP3B | chr22 | 22316911 | C | T | NM_003935 | p.472R>Q | missense | Schizophrenia | DNM:Xu_2012 |
 eQTL annotation
eQTL annotation
| SNP ID | Chromosome | Position | eGene | Gene Entrez ID | pvalue | qvalue | TSS distance | eQTL type |
|---|---|---|---|---|---|---|---|---|
| rs2283797 | 22 | 22315312 | TOP3B | ENSG00000100038.15 | 9.27199E-7 | 0.02 | 21901 | gtex_brain_ba24 |
Section II. Transcriptome annotation
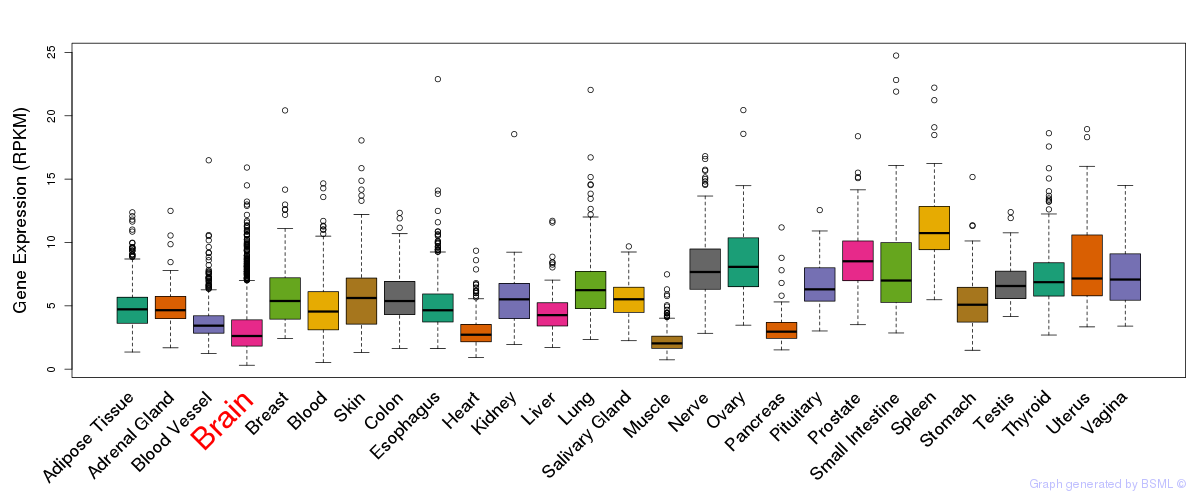
General gene expression (GTEx)
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
Top co-expressed genes in brain regions
| Top 10 positively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| ACTR3 | 0.94 | 0.95 |
| PSPC1 | 0.93 | 0.94 |
| CCT5 | 0.93 | 0.93 |
| GSPT2 | 0.92 | 0.94 |
| ARF4 | 0.92 | 0.92 |
| PLAA | 0.92 | 0.92 |
| RAB10 | 0.92 | 0.92 |
| YWHAQ | 0.92 | 0.93 |
| ARMCX5 | 0.92 | 0.92 |
| GLOD4 | 0.92 | 0.92 |
| Top 10 negatively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| MT-CO2 | -0.81 | -0.86 |
| AF347015.33 | -0.79 | -0.85 |
| FXYD1 | -0.78 | -0.84 |
| AF347015.31 | -0.78 | -0.84 |
| AF347015.8 | -0.78 | -0.85 |
| AF347015.27 | -0.76 | -0.84 |
| MT-CYB | -0.76 | -0.83 |
| AF347015.21 | -0.75 | -0.84 |
| AF347015.2 | -0.75 | -0.84 |
| AF347015.15 | -0.74 | -0.84 |
Section III. Gene Ontology annotation
| Molecular function | GO term | Evidence | Neuro keywords | PubMed ID |
|---|---|---|---|---|
| GO:0003917 | DNA topoisomerase type I activity | IEA | - | |
| GO:0005515 | protein binding | IPI | 15231747 | |
| Biological process | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0006268 | DNA unwinding during replication | IEA | - | |
| GO:0006265 | DNA topological change | IEA | - | |
| GO:0007059 | chromosome segregation | IEA | - | |
| Cellular component | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0000793 | condensed chromosome | IEA | - | |
| GO:0005634 | nucleus | TAS | 9786843 |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| KEGG HOMOLOGOUS RECOMBINATION | 28 | 17 | All SZGR 2.0 genes in this pathway |
| GAL LEUKEMIC STEM CELL UP | 133 | 78 | All SZGR 2.0 genes in this pathway |
| CONCANNON APOPTOSIS BY EPOXOMICIN UP | 239 | 157 | All SZGR 2.0 genes in this pathway |
| PUJANA BRCA1 PCC NETWORK | 1652 | 1023 | All SZGR 2.0 genes in this pathway |
| KAUFFMANN DNA REPAIR GENES | 230 | 137 | All SZGR 2.0 genes in this pathway |
| GNATENKO PLATELET SIGNATURE | 48 | 28 | All SZGR 2.0 genes in this pathway |
| PENG RAPAMYCIN RESPONSE UP | 203 | 130 | All SZGR 2.0 genes in this pathway |
| FAELT B CLL WITH VH3 21 UP | 44 | 30 | All SZGR 2.0 genes in this pathway |
| LEE CALORIE RESTRICTION NEOCORTEX DN | 88 | 58 | All SZGR 2.0 genes in this pathway |
| HELLER HDAC TARGETS UP | 317 | 208 | All SZGR 2.0 genes in this pathway |
| MATZUK MEIOTIC AND DNA REPAIR | 39 | 26 | All SZGR 2.0 genes in this pathway |
| RAO BOUND BY SALL4 ISOFORM B | 517 | 302 | All SZGR 2.0 genes in this pathway |