Gene Page: MAP3K14
Summary ?
| GeneID | 9020 |
| Symbol | MAP3K14 |
| Synonyms | FTDCR1B|HS|HSNIK|NIK |
| Description | mitogen-activated protein kinase kinase kinase 14 |
| Reference | MIM:604655|HGNC:HGNC:6853|Ensembl:ENSG00000006062|HPRD:05230|Vega:OTTHUMG00000180364 |
| Gene type | protein-coding |
| Map location | 17q21 |
| Pascal p-value | 0.653 |
| Sherlock p-value | 0.025 |
| eGene | Myers' cis & trans |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CV:PGCnp | Genome-wide Association Study | GWAS | |
| Network | Shortest path distance of core genes in the Human protein-protein interaction network | Contribution to shortest path in PPI network: 0.1134 |
Section I. Genetics and epigenetics annotation
 eQTL annotation
eQTL annotation
| SNP ID | Chromosome | Position | eGene | Gene Entrez ID | pvalue | qvalue | TSS distance | eQTL type |
|---|---|---|---|---|---|---|---|---|
| rs5988076 | chrX | 113894988 | MAP3K14 | 9020 | 0.14 | trans | ||
| rs4145870 | chrX | 114031674 | MAP3K14 | 9020 | 0.1 | trans | ||
| rs5945997 | chrX | 114039444 | MAP3K14 | 9020 | 0.1 | trans | ||
| rs5946197 | 0 | MAP3K14 | 9020 | 0.05 | trans | |||
| rs4911874 | chrX | 114087957 | MAP3K14 | 9020 | 0.13 | trans | ||
| rs5946229 | chrX | 114146933 | MAP3K14 | 9020 | 0.07 | trans | ||
| rs6644132 | chrX | 114153066 | MAP3K14 | 9020 | 0.03 | trans | ||
| rs5946239 | chrX | 114163119 | MAP3K14 | 9020 | 0.03 | trans |
Section II. Transcriptome annotation
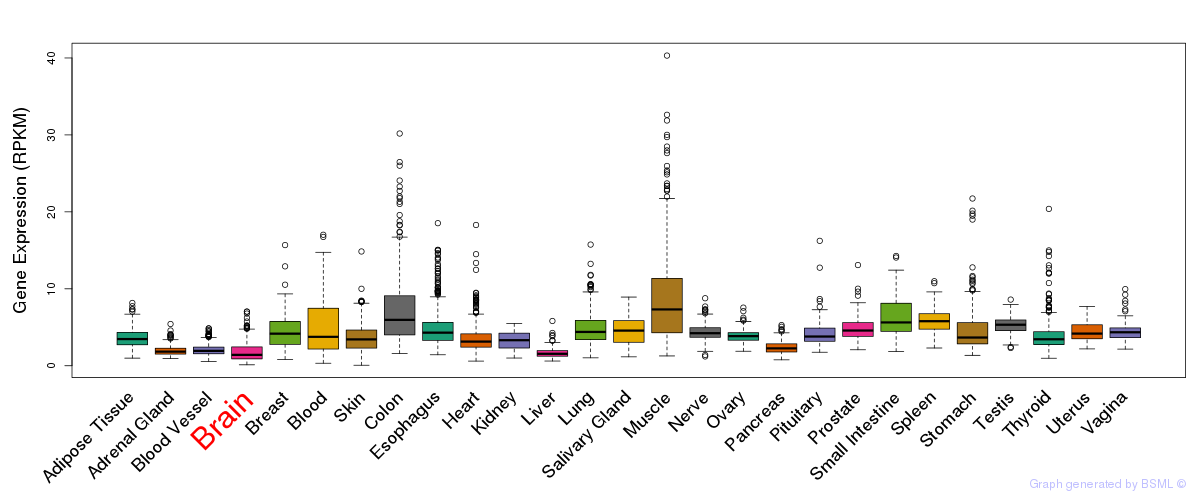
General gene expression (GTEx)
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
Top co-expressed genes in brain regions
| Top 10 positively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| SLC2A6 | 0.89 | 0.89 |
| CHAC1 | 0.86 | 0.89 |
| SLC27A4 | 0.86 | 0.85 |
| SEZ6L2 | 0.85 | 0.88 |
| CHPF | 0.85 | 0.87 |
| PPP2R5B | 0.84 | 0.85 |
| LENG4 | 0.83 | 0.88 |
| ATP13A2 | 0.83 | 0.82 |
| ZDHHC14 | 0.83 | 0.85 |
| SLC45A1 | 0.83 | 0.84 |
| Top 10 negatively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| NSBP1 | -0.48 | -0.51 |
| AF347015.21 | -0.45 | -0.33 |
| RAB13 | -0.45 | -0.54 |
| AF347015.18 | -0.45 | -0.36 |
| AF347015.8 | -0.43 | -0.32 |
| MT-ATP8 | -0.43 | -0.33 |
| AP002478.3 | -0.43 | -0.42 |
| AF347015.2 | -0.42 | -0.28 |
| ANP32B | -0.41 | -0.46 |
| AF347015.31 | -0.40 | -0.31 |
Section III. Gene Ontology annotation
| Molecular function | GO term | Evidence | Neuro keywords | PubMed ID |
|---|---|---|---|---|
| GO:0000166 | nucleotide binding | IEA | - | |
| GO:0005515 | protein binding | IPI | 9520446 |10094049 |10733566 |11035039 |12874243 |16286467 |18022362 | |
| GO:0005524 | ATP binding | IEA | - | |
| GO:0004672 | protein kinase activity | IEA | - | |
| GO:0004674 | protein serine/threonine kinase activity | IEA | - | |
| GO:0004709 | MAP kinase kinase kinase activity | IEA | - | |
| GO:0016740 | transferase activity | IEA | - | |
| Biological process | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0006468 | protein amino acid phosphorylation | IEA | - | |
| Cellular component | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0005737 | cytoplasm | IEA | - |
Section IV. Protein-protein interaction annotation
| Interactors | Aliases B | Official full name B | Experimental | Source | PubMed ID |
|---|---|---|---|---|---|
| ATP5A1 | ATP5A | ATP5AL2 | ATPM | MOM2 | OMR | ORM | hATP1 | ATP synthase, H+ transporting, mitochondrial F1 complex, alpha subunit 1, cardiac muscle | - | HPRD | 14743216 |
| CASP10 | ALPS2 | FLICE2 | MCH4 | caspase 10, apoptosis-related cysteine peptidase | Affinity Capture-Western | BioGRID | 11002417 |
| CASP3 | CPP32 | CPP32B | SCA-1 | caspase 3, apoptosis-related cysteine peptidase | Affinity Capture-Western | BioGRID | 11002417 |
| CASP8 | ALPS2B | CAP4 | FLICE | FLJ17672 | MACH | MCH5 | MGC78473 | caspase 8, apoptosis-related cysteine peptidase | - | HPRD,BioGRID | 11002417 |
| CFLAR | CASH | CASP8AP1 | CLARP | Casper | FLAME | FLAME-1 | FLAME1 | FLIP | I-FLICE | MRIT | c-FLIP | c-FLIPL | c-FLIPR | c-FLIPS | CASP8 and FADD-like apoptosis regulator | Affinity Capture-Western | BioGRID | 11002417 |
| CHUK | IKBKA | IKK-alpha | IKK1 | IKKA | NFKBIKA | TCF16 | conserved helix-loop-helix ubiquitous kinase | - | HPRD,BioGRID | 9244310 |
| CHUK | IKBKA | IKK-alpha | IKK1 | IKKA | NFKBIKA | TCF16 | conserved helix-loop-helix ubiquitous kinase | Ikk-alpha interacts with NIK. This interaction was modeled on a demonstrated interaction between human IKK-alpha and NIK from an unspecified species. | BIND | 9346485 |
| CSDA | CSDA1 | DBPA | ZONAB | cold shock domain protein A | - | HPRD | 14743216 |
| DHX9 | DDX9 | LKP | NDHII | RHA | DEAH (Asp-Glu-Ala-His) box polypeptide 9 | - | HPRD | 14743216 |
| EGFR | ERBB | ERBB1 | HER1 | PIG61 | mENA | epidermal growth factor receptor (erythroblastic leukemia viral (v-erb-b) oncogene homolog, avian) | - | HPRD,BioGRID | 11116146 |
| EXOSC10 | PM-Scl | PM/Scl-100 | PMSCL | PMSCL2 | RRP6 | Rrp6p | p2 | p3 | p4 | exosome component 10 | - | HPRD | 14743216 |
| GLUD1 | GDH | GDH1 | GLUD | MGC132003 | glutamate dehydrogenase 1 | - | HPRD | 14743216 |
| GRWD1 | GRWD | KIAA1942 | RRB1 | WDR28 | glutamate-rich WD repeat containing 1 | - | HPRD | 14743216 |
| HNRNPA1 | HNRPA1 | MGC102835 | heterogeneous nuclear ribonucleoprotein A1 | - | HPRD | 14743216 |
| HNRNPM | DKFZp547H118 | HNRNPM4 | HNRPM | HNRPM4 | HTGR1 | NAGR1 | heterogeneous nuclear ribonucleoprotein M | - | HPRD | 14743216 |
| HSPA8 | HSC54 | HSC70 | HSC71 | HSP71 | HSP73 | HSPA10 | LAP1 | MGC131511 | MGC29929 | NIP71 | heat shock 70kDa protein 8 | - | HPRD | 14743216 |
| IKBKAP | DKFZp781H1425 | DYS | ELP1 | FD | FLJ12497 | IKAP | IKI3 | TOT1 | inhibitor of kappa light polypeptide gene enhancer in B-cells, kinase complex-associated protein | - | HPRD,BioGRID | 9751059 |
| IKBKAP | DKFZp781H1425 | DYS | ELP1 | FD | FLJ12497 | IKAP | IKI3 | TOT1 | inhibitor of kappa light polypeptide gene enhancer in B-cells, kinase complex-associated protein | IKAP interacts with NIK. | BIND | 9751059 |
| IKBKB | FLJ40509 | IKK-beta | IKK2 | IKKB | MGC131801 | NFKBIKB | inhibitor of kappa light polypeptide gene enhancer in B-cells, kinase beta | IKK-beta interacts with NIK. This interaction was modeled on a demonstrated interaction between human IKK-beta and NIK from an unspecified species. | BIND | 9346485 |
| IKBKB | FLJ40509 | IKK-beta | IKK2 | IKKB | MGC131801 | NFKBIKB | inhibitor of kappa light polypeptide gene enhancer in B-cells, kinase beta | - | HPRD,BioGRID | 9346485 |
| IKBKG | AMCBX1 | FIP-3 | FIP3 | Fip3p | IKK-gamma | IP | IP1 | IP2 | IPD2 | NEMO | inhibitor of kappa light polypeptide gene enhancer in B-cells, kinase gamma | - | HPRD,BioGRID | 9927690 |
| MAP3K14 | FTDCR1B | HS | HSNIK | NIK | mitogen-activated protein kinase kinase kinase 14 | - | HPRD,BioGRID | 10887201 |
| MAP3K7 | TAK1 | TGF1a | mitogen-activated protein kinase kinase kinase 7 | - | HPRD,BioGRID | 10094049 |
| MAP3K8 | COT | EST | ESTF | FLJ10486 | TPL2 | Tpl-2 | c-COT | mitogen-activated protein kinase kinase kinase 8 | - | HPRD,BioGRID | 10072079 |
| NFKB2 | LYT-10 | LYT10 | nuclear factor of kappa light polypeptide gene enhancer in B-cells 2 (p49/p100) | - | HPRD,BioGRID | 11239468 |
| NFKBIA | IKBA | MAD-3 | NFKBI | nuclear factor of kappa light polypeptide gene enhancer in B-cells inhibitor, alpha | Reconstituted Complex | BioGRID | 9751059 |
| NPM1 | B23 | MGC104254 | NPM | nucleophosmin (nucleolar phosphoprotein B23, numatrin) | - | HPRD | 14743216 |
| PELI3 | MGC35521 | pellino homolog 3 (Drosophila) | - | HPRD | 12874243 |
| RIPK1 | FLJ39204 | RIP | RIP1 | receptor (TNFRSF)-interacting serine-threonine kinase 1 | - | HPRD,BioGRID | 11116146 |
| RPL10A | Csa-19 | NEDD6 | ribosomal protein L10a | - | HPRD | 14743216 |
| RPL18 | - | ribosomal protein L18 | - | HPRD | 14743216 |
| RPL24 | - | ribosomal protein L24 | - | HPRD | 14743216 |
| RPL3 | MGC104284 | TARBP-B | ribosomal protein L3 | - | HPRD | 14743216 |
| RPL32 | PP9932 | ribosomal protein L32 | - | HPRD | 14743216 |
| RPL35 | - | ribosomal protein L35 | - | HPRD | 14743216 |
| RPL8 | - | ribosomal protein L8 | - | HPRD | 14743216 |
| RPS3 | FLJ26283 | FLJ27450 | MGC87870 | ribosomal protein S3 | - | HPRD | 14743216 |
| RRP12 | DKFZp762P1116 | FLJ20231 | KIAA0690 | ribosomal RNA processing 12 homolog (S. cerevisiae) | - | HPRD | 14743216 |
| RUVBL2 | CGI-46 | ECP51 | INO80J | REPTIN | RVB2 | TIH2 | TIP48 | TIP49B | RuvB-like 2 (E. coli) | - | HPRD | 14743216 |
| TRAF1 | EBI6 | MGC:10353 | TNF receptor-associated factor 1 | - | HPRD,BioGRID | 11278268 |
| TRAF1 | EBI6 | MGC:10353 | TNF receptor-associated factor 1 | NIK interacts with TRAF1. This interaction was modeled on a demonstrated interaction between human NIK and TRAF1 from an unspecified species. | BIND | 9275204 |
| TRAF2 | MGC:45012 | TRAP | TRAP3 | TNF receptor-associated factor 2 | - | HPRD,BioGRID | 9020361 |11278268 |
| TRAF2 | MGC:45012 | TRAP | TRAP3 | TNF receptor-associated factor 2 | NIK interacts with TRAF2. This interaction was modeled on a demonstrated interaction between human NIK and TRAF2 from an unspecified species. | BIND | 9275204 |
| TRAF3 | CAP-1 | CD40bp | CRAF1 | LAP1 | TNF receptor-associated factor 3 | - | HPRD,BioGRID | 11278268 |
| TRAF3 | CAP-1 | CD40bp | CRAF1 | LAP1 | TNF receptor-associated factor 3 | NIK interacts with TRAF3. This interaction was modeled on a demonstrated interaction between human NIK and TRAF3 from an unspecified species. | BIND | 9275204 |
| TRAF5 | MGC:39780 | RNF84 | TNF receptor-associated factor 5 | NIK interacts with TRAF5. This interaction was modeled on a demonstrated interaction between human NIK and TRAF5 from an unspecified species. | BIND | 9275204 |
| TRAF6 | MGC:3310 | RNF85 | TNF receptor-associated factor 6 | - | HPRD,BioGRID | 11278268 |
| TRAF6 | MGC:3310 | RNF85 | TNF receptor-associated factor 6 | NIK interacts with TRAF6. This interaction was modeled on a demonstrated interaction between human NIK and TRAF6 from an unspecified species. | BIND | 9275204 |
| TUBA3C | TUBA2 | bA408E5.3 | tubulin, alpha 3c | - | HPRD | 14743216 |
| TUBB2B | DKFZp566F223 | FLJ98847 | MGC8685 | TUBB-PARALOG | bA506K6.1 | tubulin, beta 2B | - | HPRD | 14743216 |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| KEGG MAPK SIGNALING PATHWAY | 267 | 205 | All SZGR 2.0 genes in this pathway |
| KEGG APOPTOSIS | 88 | 62 | All SZGR 2.0 genes in this pathway |
| KEGG T CELL RECEPTOR SIGNALING PATHWAY | 108 | 89 | All SZGR 2.0 genes in this pathway |
| KEGG INTESTINAL IMMUNE NETWORK FOR IGA PRODUCTION | 48 | 36 | All SZGR 2.0 genes in this pathway |
| KEGG EPITHELIAL CELL SIGNALING IN HELICOBACTER PYLORI INFECTION | 68 | 44 | All SZGR 2.0 genes in this pathway |
| BIOCARTA CD40 PATHWAY | 15 | 13 | All SZGR 2.0 genes in this pathway |
| BIOCARTA RNA PATHWAY | 10 | 9 | All SZGR 2.0 genes in this pathway |
| BIOCARTA HIVNEF PATHWAY | 58 | 43 | All SZGR 2.0 genes in this pathway |
| BIOCARTA DEATH PATHWAY | 33 | 24 | All SZGR 2.0 genes in this pathway |
| BIOCARTA KERATINOCYTE PATHWAY | 46 | 38 | All SZGR 2.0 genes in this pathway |
| BIOCARTA MAPK PATHWAY | 87 | 68 | All SZGR 2.0 genes in this pathway |
| BIOCARTA NTHI PATHWAY | 24 | 20 | All SZGR 2.0 genes in this pathway |
| BIOCARTA NFKB PATHWAY | 23 | 19 | All SZGR 2.0 genes in this pathway |
| BIOCARTA IL1R PATHWAY | 33 | 29 | All SZGR 2.0 genes in this pathway |
| BIOCARTA TALL1 PATHWAY | 15 | 15 | All SZGR 2.0 genes in this pathway |
| BIOCARTA STRESS PATHWAY | 25 | 18 | All SZGR 2.0 genes in this pathway |
| BIOCARTA TNFR2 PATHWAY | 18 | 15 | All SZGR 2.0 genes in this pathway |
| BIOCARTA TOLL PATHWAY | 37 | 31 | All SZGR 2.0 genes in this pathway |
| PID TCR PATHWAY | 66 | 51 | All SZGR 2.0 genes in this pathway |
| PID CD40 PATHWAY | 31 | 26 | All SZGR 2.0 genes in this pathway |
| PID AVB3 OPN PATHWAY | 31 | 29 | All SZGR 2.0 genes in this pathway |
| PID CD8 TCR PATHWAY | 53 | 42 | All SZGR 2.0 genes in this pathway |
| PID HIV NEF PATHWAY | 35 | 26 | All SZGR 2.0 genes in this pathway |
| REACTOME CD28 CO STIMULATION | 32 | 26 | All SZGR 2.0 genes in this pathway |
| REACTOME COSTIMULATION BY THE CD28 FAMILY | 63 | 48 | All SZGR 2.0 genes in this pathway |
| REACTOME CD28 DEPENDENT PI3K AKT SIGNALING | 22 | 18 | All SZGR 2.0 genes in this pathway |
| REACTOME IMMUNE SYSTEM | 933 | 616 | All SZGR 2.0 genes in this pathway |
| REACTOME ADAPTIVE IMMUNE SYSTEM | 539 | 350 | All SZGR 2.0 genes in this pathway |
| ZHONG RESPONSE TO AZACITIDINE AND TSA UP | 183 | 119 | All SZGR 2.0 genes in this pathway |
| HOOI ST7 TARGETS UP | 94 | 57 | All SZGR 2.0 genes in this pathway |
| NAGASHIMA NRG1 SIGNALING UP | 176 | 123 | All SZGR 2.0 genes in this pathway |
| KOKKINAKIS METHIONINE DEPRIVATION 48HR DN | 64 | 39 | All SZGR 2.0 genes in this pathway |
| KOKKINAKIS METHIONINE DEPRIVATION 96HR DN | 75 | 50 | All SZGR 2.0 genes in this pathway |
| LINDGREN BLADDER CANCER CLUSTER 1 DN | 378 | 231 | All SZGR 2.0 genes in this pathway |
| CONCANNON APOPTOSIS BY EPOXOMICIN UP | 239 | 157 | All SZGR 2.0 genes in this pathway |
| PEREZ TP53 TARGETS | 1174 | 695 | All SZGR 2.0 genes in this pathway |
| SCHEIDEREIT IKK INTERACTING PROTEINS | 58 | 45 | All SZGR 2.0 genes in this pathway |
| DACOSTA UV RESPONSE VIA ERCC3 DN | 855 | 609 | All SZGR 2.0 genes in this pathway |
| NUYTTEN NIPP1 TARGETS DN | 848 | 527 | All SZGR 2.0 genes in this pathway |
| NUYTTEN EZH2 TARGETS UP | 1037 | 673 | All SZGR 2.0 genes in this pathway |
| BUYTAERT PHOTODYNAMIC THERAPY STRESS UP | 811 | 508 | All SZGR 2.0 genes in this pathway |
| KOYAMA SEMA3B TARGETS DN | 411 | 249 | All SZGR 2.0 genes in this pathway |
| AMIT EGF RESPONSE 120 HELA | 69 | 47 | All SZGR 2.0 genes in this pathway |
| AMIT EGF RESPONSE 60 MCF10A | 39 | 24 | All SZGR 2.0 genes in this pathway |
| AMIT SERUM RESPONSE 120 MCF10A | 65 | 44 | All SZGR 2.0 genes in this pathway |
| STANELLE E2F1 TARGETS | 29 | 20 | All SZGR 2.0 genes in this pathway |
| NATSUME RESPONSE TO INTERFERON BETA UP | 71 | 49 | All SZGR 2.0 genes in this pathway |
| MURAKAMI UV RESPONSE 6HR DN | 21 | 13 | All SZGR 2.0 genes in this pathway |
| BLALOCK ALZHEIMERS DISEASE UP | 1691 | 1088 | All SZGR 2.0 genes in this pathway |
| MURAKAMI UV RESPONSE 24HR | 20 | 13 | All SZGR 2.0 genes in this pathway |
| MARSON BOUND BY FOXP3 UNSTIMULATED | 1229 | 713 | All SZGR 2.0 genes in this pathway |
| IWANAGA CARCINOGENESIS BY KRAS DN | 120 | 81 | All SZGR 2.0 genes in this pathway |
| MITSIADES RESPONSE TO APLIDIN UP | 439 | 257 | All SZGR 2.0 genes in this pathway |
| FIRESTEIN CTNNB1 PATHWAY AND PROLIFERATION | 9 | 5 | All SZGR 2.0 genes in this pathway |
| FIRESTEIN CTNNB1 PATHWAY | 33 | 23 | All SZGR 2.0 genes in this pathway |
| FIRESTEIN PROLIFERATION | 175 | 125 | All SZGR 2.0 genes in this pathway |
| LINDSTEDT DENDRITIC CELL MATURATION B | 53 | 36 | All SZGR 2.0 genes in this pathway |
| RUTELLA RESPONSE TO CSF2RB AND IL4 UP | 338 | 225 | All SZGR 2.0 genes in this pathway |
| RUTELLA RESPONSE TO HGF DN | 235 | 144 | All SZGR 2.0 genes in this pathway |
| RUTELLA RESPONSE TO HGF VS CSF2RB AND IL4 DN | 245 | 150 | All SZGR 2.0 genes in this pathway |
| CAIRO HEPATOBLASTOMA DN | 267 | 160 | All SZGR 2.0 genes in this pathway |
| CAIRO HEPATOBLASTOMA CLASSES DN | 210 | 141 | All SZGR 2.0 genes in this pathway |
| MURAKAMI UV RESPONSE 1HR UP | 17 | 10 | All SZGR 2.0 genes in this pathway |
| DUTERTRE ESTRADIOL RESPONSE 24HR DN | 505 | 328 | All SZGR 2.0 genes in this pathway |
| KOINUMA TARGETS OF SMAD2 OR SMAD3 | 824 | 528 | All SZGR 2.0 genes in this pathway |
| PLASARI TGFB1 TARGETS 1HR UP | 34 | 26 | All SZGR 2.0 genes in this pathway |
| KRIEG HYPOXIA VIA KDM3A | 53 | 34 | All SZGR 2.0 genes in this pathway |
| ZWANG CLASS 1 TRANSIENTLY INDUCED BY EGF | 516 | 308 | All SZGR 2.0 genes in this pathway |
| ZWANG TRANSIENTLY UP BY 2ND EGF PULSE ONLY | 1725 | 838 | All SZGR 2.0 genes in this pathway |
Section VI. microRNA annotation
| miRNA family | Target position | miRNA ID | miRNA seq | ||
|---|---|---|---|---|---|
| UTR start | UTR end | Match method | |||
| miR-130/301 | 1013 | 1019 | m8 | hsa-miR-130abrain | CAGUGCAAUGUUAAAAGGGCAU |
| hsa-miR-301 | CAGUGCAAUAGUAUUGUCAAAGC | ||||
| hsa-miR-130bbrain | CAGUGCAAUGAUGAAAGGGCAU | ||||
| hsa-miR-454-3p | UAGUGCAAUAUUGCUUAUAGGGUUU | ||||
| miR-155 | 1247 | 1254 | 1A,m8 | hsa-miR-155 | UUAAUGCUAAUCGUGAUAGGGG |
| miR-17-5p/20/93.mr/106/519.d | 1034 | 1040 | m8 | hsa-miR-17-5p | CAAAGUGCUUACAGUGCAGGUAGU |
| hsa-miR-20abrain | UAAAGUGCUUAUAGUGCAGGUAG | ||||
| hsa-miR-106a | AAAAGUGCUUACAGUGCAGGUAGC | ||||
| hsa-miR-106bSZ | UAAAGUGCUGACAGUGCAGAU | ||||
| hsa-miR-20bSZ | CAAAGUGCUCAUAGUGCAGGUAG | ||||
| hsa-miR-519d | CAAAGUGCCUCCCUUUAGAGUGU | ||||
| hsa-miR-17-5p | CAAAGUGCUUACAGUGCAGGUAGU | ||||
| hsa-miR-20abrain | UAAAGUGCUUAUAGUGCAGGUAG | ||||
| hsa-miR-106a | AAAAGUGCUUACAGUGCAGGUAGC | ||||
| hsa-miR-106bSZ | UAAAGUGCUGACAGUGCAGAU | ||||
| hsa-miR-20bSZ | CAAAGUGCUCAUAGUGCAGGUAG | ||||
| hsa-miR-519d | CAAAGUGCCUCCCUUUAGAGUGU | ||||
| miR-19 | 1012 | 1018 | m8 | hsa-miR-19a | UGUGCAAAUCUAUGCAAAACUGA |
| hsa-miR-19b | UGUGCAAAUCCAUGCAAAACUGA | ||||
| miR-27 | 1260 | 1266 | 1A | hsa-miR-27abrain | UUCACAGUGGCUAAGUUCCGC |
| hsa-miR-27bbrain | UUCACAGUGGCUAAGUUCUGC | ||||
| miR-93.hd/291-3p/294/295/302/372/373/520 | 1033 | 1039 | m8 | hsa-miR-93brain | AAAGUGCUGUUCGUGCAGGUAG |
| hsa-miR-302a | UAAGUGCUUCCAUGUUUUGGUGA | ||||
| hsa-miR-302b | UAAGUGCUUCCAUGUUUUAGUAG | ||||
| hsa-miR-302c | UAAGUGCUUCCAUGUUUCAGUGG | ||||
| hsa-miR-302d | UAAGUGCUUCCAUGUUUGAGUGU | ||||
| hsa-miR-372 | AAAGUGCUGCGACAUUUGAGCGU | ||||
| hsa-miR-373 | GAAGUGCUUCGAUUUUGGGGUGU | ||||
| hsa-miR-520e | AAAGUGCUUCCUUUUUGAGGG | ||||
| hsa-miR-520a | AAAGUGCUUCCCUUUGGACUGU | ||||
| hsa-miR-520b | AAAGUGCUUCCUUUUAGAGGG | ||||
| hsa-miR-520c | AAAGUGCUUCCUUUUAGAGGGUU | ||||
| hsa-miR-520d | AAAGUGCUUCUCUUUGGUGGGUU | ||||
- SZ: miRNAs which differentially expressed in brain cortex of schizophrenia patients comparing with control samples using microarray. Click here to see the list of SZ related miRNAs.
- Brain: miRNAs which are expressed in brain based on miRNA microarray expression studies. Click here to see the list of brain related miRNAs.