Gene Page: ASH2L
Summary ?
| GeneID | 9070 |
| Symbol | ASH2L |
| Synonyms | ASH2|ASH2L1|ASH2L2|Bre2 |
| Description | ash2 (absent, small, or homeotic)-like (Drosophila) |
| Reference | MIM:604782|HGNC:HGNC:744|Ensembl:ENSG00000129691|HPRD:05309|Vega:OTTHUMG00000164016 |
| Gene type | protein-coding |
| Map location | 8p11.2 |
| Pascal p-value | 1.842E-5 |
| Sherlock p-value | 0.241 |
| Fetal beta | -0.083 |
| DMG | 1 (# studies) |
| eGene | Myers' cis & trans |
| Support | Chromatin Remodeling Genes |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CV:PGCnp | Genome-wide Association Study | GWAS | |
| DMG:Wockner_2014 | Genome-wide DNA methylation analysis | This dataset includes 4641 differentially methylated probes corresponding to 2929 unique genes between schizophrenia patients (n=24) and controls (n=24). | 1 |
| PMID:cooccur | High-throughput literature-search | Systematic search in PubMed for genes co-occurring with SCZ keywords. A total of 3027 genes were included. | |
| Literature | High-throughput literature-search | Co-occurance with Schizophrenia keywords: schizophrenia,schizophrenias | Click to show details |
Section I. Genetics and epigenetics annotation
 Differentially methylated gene
Differentially methylated gene
| Probe | Chromosome | Position | Nearest gene | P (dis) | Beta (dis) | FDR (dis) | Study |
|---|---|---|---|---|---|---|---|
| cg03024183 | 8 | 37963317 | ASH2L | 2.26E-7 | -0.311 | 0.005 | DMG:Wockner_2014 |
 eQTL annotation
eQTL annotation
| SNP ID | Chromosome | Position | eGene | Gene Entrez ID | pvalue | qvalue | TSS distance | eQTL type |
|---|---|---|---|---|---|---|---|---|
| rs7132043 | chr12 | 80968399 | ASH2L | 9070 | 0.14 | trans |
Section II. Transcriptome annotation
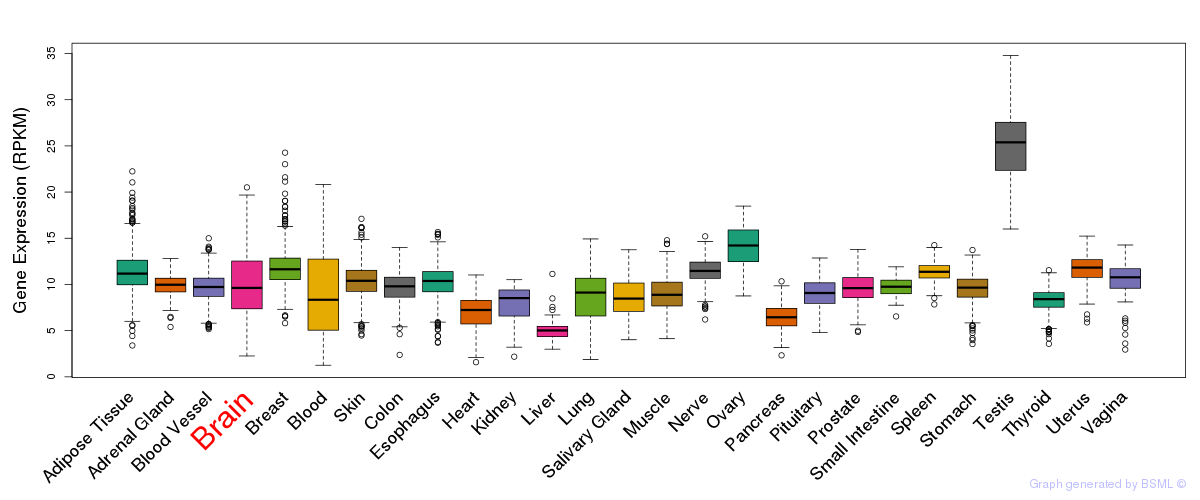
General gene expression (GTEx)
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
Top co-expressed genes in brain regions
| Top 10 positively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| ATRNL1 | 0.90 | 0.87 |
| CACNB4 | 0.89 | 0.82 |
| FAM126B | 0.89 | 0.87 |
| GABRB2 | 0.88 | 0.75 |
| SORL1 | 0.87 | 0.80 |
| UBR3 | 0.87 | 0.88 |
| TTC39B | 0.87 | 0.78 |
| ARFGEF2 | 0.86 | 0.83 |
| CHM | 0.86 | 0.84 |
| CPEB3 | 0.86 | 0.83 |
| Top 10 negatively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| EFEMP2 | -0.50 | -0.60 |
| RAB34 | -0.48 | -0.64 |
| GATSL3 | -0.44 | -0.55 |
| RHOC | -0.43 | -0.61 |
| DBI | -0.43 | -0.60 |
| NKAIN4 | -0.41 | -0.55 |
| C19orf36 | -0.41 | -0.53 |
| NUDT8 | -0.40 | -0.49 |
| C1orf61 | -0.40 | -0.67 |
| VEGFB | -0.39 | -0.49 |
Section IV. Protein-protein interaction annotation
| Interactors | Aliases B | Official full name B | Experimental | Source | PubMed ID |
|---|---|---|---|---|---|
| ASH2L | ASH2 | ASH2L1 | ASH2L2 | Bre2 | ash2 (absent, small, or homeotic)-like (Drosophila) | Reconstituted Complex | BioGRID | 15199122 |
| DPY30 | HDPY-30 | Saf19 | dpy-30 homolog (C. elegans) | Two-hybrid | BioGRID | 16189514 |
| HCFC1 | CFF | HCF-1 | HCF1 | HFC1 | MGC70925 | VCAF | host cell factor C1 (VP16-accessory protein) | Reconstituted Complex | BioGRID | 15199122 |
| HCFC2 | FLJ94012 | HCF-2 | HCF2 | host cell factor C2 | Reconstituted Complex | BioGRID | 15199122 |
| HIST3H3 | H3.4 | H3/g | H3FT | H3t | MGC126886 | MGC126888 | histone cluster 3, H3 | Reconstituted Complex | BioGRID | 12482968 |
| MEN1 | MEAI | SCG2 | multiple endocrine neoplasia I | Reconstituted Complex | BioGRID | 15199122 |
| MLL | ALL-1 | CXXC7 | FLJ11783 | HRX | HTRX1 | KMT2A | MLL/GAS7 | MLL1A | TET1-MLL | TRX1 | myeloid/lymphoid or mixed-lineage leukemia (trithorax homolog, Drosophila) | Affinity Capture-Western Co-purification | BioGRID | 15199122 |
| MLL3 | DKFZp686C08112 | FLJ12625 | FLJ38309 | HALR | KIAA1506 | KMT2C | MGC119851 | MGC119852 | MGC119853 | myeloid/lymphoid or mixed-lineage leukemia 3 | Reconstituted Complex | BioGRID | 12482968 |
| RBBP5 | RBQ3 | SWD1 | retinoblastoma binding protein 5 | Reconstituted Complex | BioGRID | 15199122 |
| UNC119 | HRG4 | unc-119 homolog (C. elegans) | Two-hybrid | BioGRID | 16169070 |
| WDR5 | BIG-3 | SWD3 | WD repeat domain 5 | Reconstituted Complex | BioGRID | 15199122 |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| HOLLMANN APOPTOSIS VIA CD40 UP | 201 | 125 | All SZGR 2.0 genes in this pathway |
| CASORELLI ACUTE PROMYELOCYTIC LEUKEMIA DN | 663 | 425 | All SZGR 2.0 genes in this pathway |
| DIAZ CHRONIC MEYLOGENOUS LEUKEMIA UP | 1382 | 904 | All SZGR 2.0 genes in this pathway |
| ENK UV RESPONSE KERATINOCYTE DN | 485 | 334 | All SZGR 2.0 genes in this pathway |
| JAERVINEN AMPLIFIED IN LARYNGEAL CANCER | 40 | 24 | All SZGR 2.0 genes in this pathway |
| SPIELMAN LYMPHOBLAST EUROPEAN VS ASIAN DN | 584 | 395 | All SZGR 2.0 genes in this pathway |
| PUJANA BRCA1 PCC NETWORK | 1652 | 1023 | All SZGR 2.0 genes in this pathway |
| BENPORATH MYC MAX TARGETS | 775 | 494 | All SZGR 2.0 genes in this pathway |
| NIKOLSKY BREAST CANCER 8P12 P11 AMPLICON | 57 | 32 | All SZGR 2.0 genes in this pathway |
| DING LUNG CANCER EXPRESSION BY COPY NUMBER | 100 | 62 | All SZGR 2.0 genes in this pathway |
| BLALOCK ALZHEIMERS DISEASE DN | 1237 | 837 | All SZGR 2.0 genes in this pathway |
| YAMAZAKI TCEB3 TARGETS DN | 215 | 132 | All SZGR 2.0 genes in this pathway |
| MCCLUNG CREB1 TARGETS UP | 100 | 72 | All SZGR 2.0 genes in this pathway |
| MONNIER POSTRADIATION TUMOR ESCAPE UP | 393 | 244 | All SZGR 2.0 genes in this pathway |
| ALONSO METASTASIS UP | 198 | 128 | All SZGR 2.0 genes in this pathway |
| WANG TUMOR INVASIVENESS UP | 374 | 247 | All SZGR 2.0 genes in this pathway |
| AGUIRRE PANCREATIC CANCER COPY NUMBER UP | 298 | 174 | All SZGR 2.0 genes in this pathway |
| MIKKELSEN PLURIPOTENT STATE UP | 11 | 8 | All SZGR 2.0 genes in this pathway |
| YAGI AML WITH INV 16 TRANSLOCATION | 422 | 277 | All SZGR 2.0 genes in this pathway |
| DANG BOUND BY MYC | 1103 | 714 | All SZGR 2.0 genes in this pathway |