Gene Page: PRPF3
Summary ?
| GeneID | 9129 |
| Symbol | PRPF3 |
| Synonyms | HPRP3|HPRP3P|PRP3|Prp3p|RP18|SNRNP90 |
| Description | pre-mRNA processing factor 3 |
| Reference | MIM:607301|HGNC:HGNC:17348|Ensembl:ENSG00000117360|HPRD:07389|Vega:OTTHUMG00000012807 |
| Gene type | protein-coding |
| Map location | 1q21.1 |
| Fetal beta | 1.117 |
| eGene | Myers' cis & trans |
| Support | Ascano FMRP targets |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CV:GWAScat | Genome-wide Association Studies | This data set includes 560 SNPs associated with schizophrenia. A total of 486 genes were mapped to these SNPs within 50kb. | |
| CV:GWASdb | Genome-wide Association Studies | GWASdb records for schizophrenia | |
| CV:PGCnp | Genome-wide Association Study | GWAS | |
| GSMA_I | Genome scan meta-analysis | Psr: 0.0235 | |
| GSMA_IIA | Genome scan meta-analysis (All samples) | Psr: 0.00814 |
Section I. Genetics and epigenetics annotation
 eQTL annotation
eQTL annotation
| SNP ID | Chromosome | Position | eGene | Gene Entrez ID | pvalue | qvalue | TSS distance | eQTL type |
|---|---|---|---|---|---|---|---|---|
| rs16955618 | chr15 | 29937543 | PRPF3 | 9129 | 0.18 | trans |
Section II. Transcriptome annotation
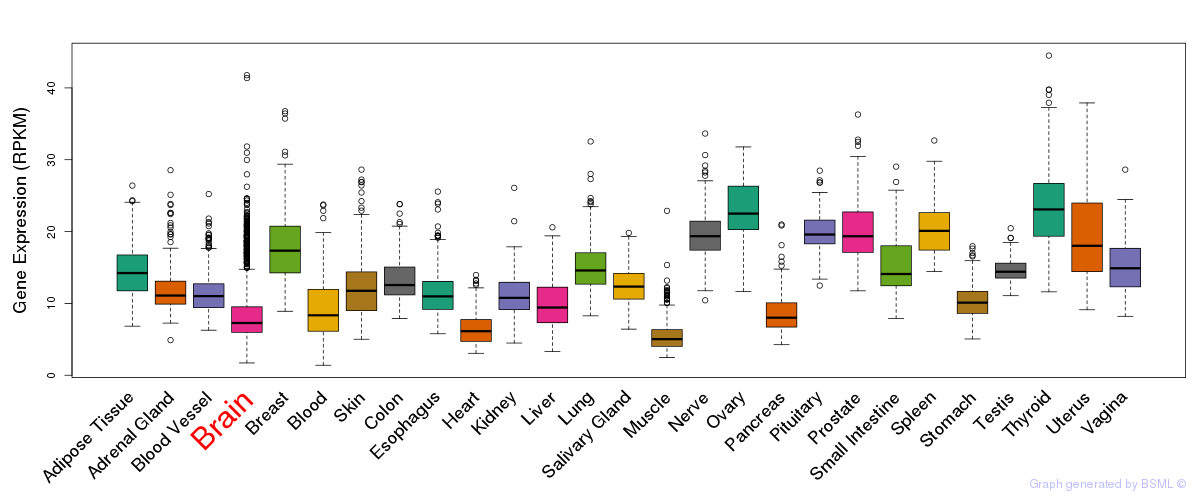
General gene expression (GTEx)
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
Top co-expressed genes in brain regions
| Top 10 positively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| VWA5B2 | 0.78 | 0.75 |
| ARHGEF4 | 0.77 | 0.74 |
| PDE4C | 0.77 | 0.75 |
| SHANK3 | 0.75 | 0.80 |
| DGKZ | 0.75 | 0.72 |
| KCNIP3 | 0.75 | 0.73 |
| SAPS1 | 0.75 | 0.72 |
| PACSIN1 | 0.74 | 0.74 |
| SYT15 | 0.74 | 0.77 |
| FMNL1 | 0.74 | 0.68 |
| Top 10 negatively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| RPS6 | -0.52 | -0.63 |
| RPL24 | -0.51 | -0.64 |
| C9orf46 | -0.51 | -0.58 |
| RPS13P2 | -0.50 | -0.59 |
| RPS18 | -0.50 | -0.60 |
| RPS3AP47 | -0.50 | -0.56 |
| RPL27 | -0.50 | -0.61 |
| RPL5 | -0.50 | -0.54 |
| AC079354.7 | -0.50 | -0.52 |
| RPS23 | -0.50 | -0.59 |
Section III. Gene Ontology annotation
| Molecular function | GO term | Evidence | Neuro keywords | PubMed ID |
|---|---|---|---|---|
| GO:0005515 | protein binding | IPI | 16189514 |17353931 | |
| GO:0031202 | RNA splicing factor activity, transesterification mechanism | TAS | 9328476 | |
| Biological process | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0000398 | nuclear mRNA splicing, via spliceosome | NAS | 9328476 | |
| GO:0008380 | RNA splicing | TAS | 9328476 | |
| GO:0007601 | visual perception | IEA | - | |
| GO:0050896 | response to stimulus | IEA | - | |
| Cellular component | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0005634 | nucleus | TAS | 9328476 | |
| GO:0005681 | spliceosome | NAS | 9328476 | |
| GO:0016607 | nuclear speck | IEA | - |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| KEGG SPLICEOSOME | 128 | 72 | All SZGR 2.0 genes in this pathway |
| BASAKI YBX1 TARGETS UP | 290 | 177 | All SZGR 2.0 genes in this pathway |
| OSMAN BLADDER CANCER DN | 406 | 230 | All SZGR 2.0 genes in this pathway |
| ENK UV RESPONSE KERATINOCYTE UP | 530 | 342 | All SZGR 2.0 genes in this pathway |
| GAUSSMANN MLL AF4 FUSION TARGETS A UP | 191 | 128 | All SZGR 2.0 genes in this pathway |
| CHIBA RESPONSE TO TSA | 50 | 31 | All SZGR 2.0 genes in this pathway |
| BROWNE HCMV INFECTION 14HR UP | 156 | 101 | All SZGR 2.0 genes in this pathway |
| MONNIER POSTRADIATION TUMOR ESCAPE UP | 393 | 244 | All SZGR 2.0 genes in this pathway |
| ACEVEDO LIVER CANCER UP | 973 | 570 | All SZGR 2.0 genes in this pathway |
| WEST ADRENOCORTICAL CARCINOMA VS ADENOMA UP | 21 | 13 | All SZGR 2.0 genes in this pathway |
| CHEN HOXA5 TARGETS 9HR UP | 223 | 132 | All SZGR 2.0 genes in this pathway |
| PILON KLF1 TARGETS DN | 1972 | 1213 | All SZGR 2.0 genes in this pathway |
| BRUINS UVC RESPONSE VIA TP53 GROUP D | 280 | 158 | All SZGR 2.0 genes in this pathway |