Gene Page: MAGI1
Summary ?
| GeneID | 9223 |
| Symbol | MAGI1 |
| Synonyms | AIP-3|AIP3|BAIAP1|BAP-1|BAP1|MAGI-1|Magi1d|TNRC19|WWP3 |
| Description | membrane associated guanylate kinase, WW and PDZ domain containing 1 |
| Reference | MIM:602625|HGNC:HGNC:946|Ensembl:ENSG00000151276|HPRD:04020|Vega:OTTHUMG00000157554 |
| Gene type | protein-coding |
| Map location | 3p14.1 |
| Pascal p-value | 0.651 |
| Sherlock p-value | 0.843 |
| Fetal beta | -1.107 |
| eGene | Putamen basal ganglia Myers' cis & trans Meta |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CV:GWASdb | Genome-wide Association Studies | GWASdb records for schizophrenia | |
| CV:PGCnp | Genome-wide Association Study | GWAS | |
| PMID:cooccur | High-throughput literature-search | Systematic search in PubMed for genes co-occurring with SCZ keywords. A total of 3027 genes were included. | |
| Association | A combined odds ratio method (Sun et al. 2008), association studies | 2 | Link to SZGene |
| Literature | High-throughput literature-search | Co-occurance with Schizophrenia keywords: schizophrenia,schizophrenic,schizophrenics,schizophrenias | Click to show details |
| GO_Annotation | Mapping neuro-related keywords to Gene Ontology annotations | Hits with neuro-related keywords: 1 |
Section I. Genetics and epigenetics annotation
 eQTL annotation
eQTL annotation
| SNP ID | Chromosome | Position | eGene | Gene Entrez ID | pvalue | qvalue | TSS distance | eQTL type |
|---|---|---|---|---|---|---|---|---|
| rs16955618 | chr15 | 29937543 | MAGI1 | 9223 | 0.18 | trans | ||
| rs12488399 | 3 | 65216032 | MAGI1 | ENSG00000151276.19 | 5.29466E-7 | 0.01 | 808193 | gtex_brain_putamen_basal |
| rs12488533 | 3 | 65216274 | MAGI1 | ENSG00000151276.19 | 3.00438E-7 | 0.01 | 807951 | gtex_brain_putamen_basal |
Section II. Transcriptome annotation
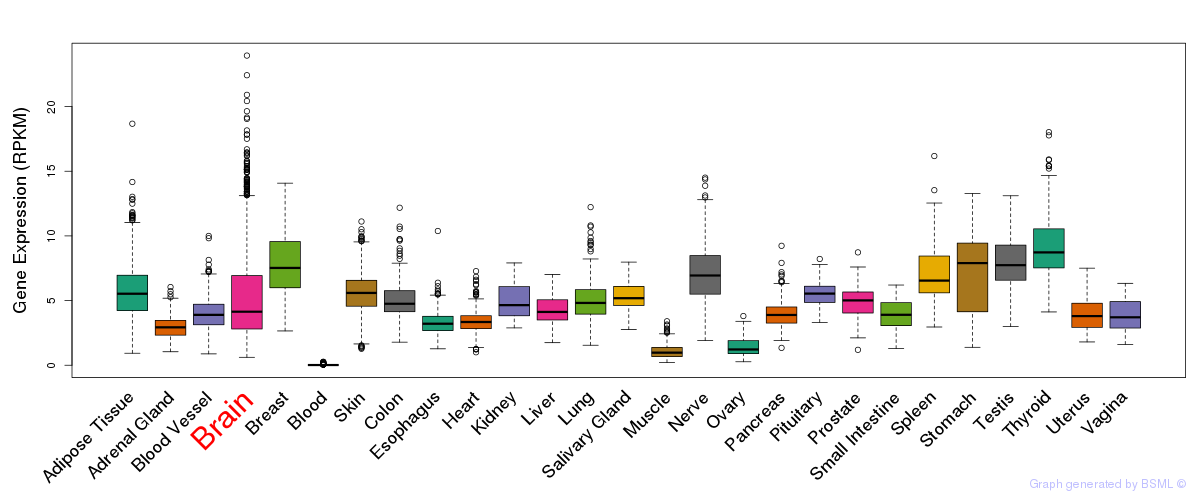
General gene expression (GTEx)
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
Top co-expressed genes in brain regions
| Top 10 positively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| DSP | 0.87 | 0.66 |
| CFH | 0.86 | 0.61 |
| OGN | 0.86 | 0.59 |
| GJB2 | 0.85 | 0.46 |
| SCARA5 | 0.85 | 0.61 |
| BNC2 | 0.83 | 0.48 |
| SIX1 | 0.81 | 0.46 |
| SIX2 | 0.79 | 0.36 |
| SLC47A1 | 0.79 | 0.60 |
| FOXC2 | 0.79 | 0.49 |
| Top 10 negatively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| HES4 | -0.24 | -0.39 |
| AC217773.2 | -0.21 | -0.30 |
| FAM128A | -0.21 | -0.34 |
| MRPL41 | -0.20 | -0.10 |
| C1orf61 | -0.20 | -0.27 |
| NUDT8 | -0.19 | -0.30 |
| NBR2 | -0.19 | -0.14 |
| C7orf55 | -0.19 | -0.26 |
| C9orf142 | -0.18 | -0.29 |
| C16orf14 | -0.18 | -0.11 |
Section III. Gene Ontology annotation
| Molecular function | GO term | Evidence | Neuro keywords | PubMed ID |
|---|---|---|---|---|
| GO:0000166 | nucleotide binding | IEA | - | |
| GO:0005524 | ATP binding | IEA | - | |
| GO:0008022 | protein C-terminus binding | TAS | 9647739 | |
| Biological process | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0007155 | cell adhesion | TAS | 9647739 | |
| GO:0006461 | protein complex assembly | NAS | 9647739 | |
| GO:0007166 | cell surface receptor linked signal transduction | TAS | 9647739 | |
| Cellular component | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0005923 | tight junction | IEA | Brain (GO term level: 10) | - |
| GO:0005886 | plasma membrane | TAS | 9647739 |
Section IV. Protein-protein interaction annotation
| Interactors | Aliases B | Official full name B | Experimental | Source | PubMed ID |
|---|---|---|---|---|---|
| ACCN3 | ASIC3 | DRASIC | SLNAC1 | TNaC1 | amiloride-sensitive cation channel 3 | - | HPRD,BioGRID | 15317815 |
| ACCN3 | ASIC3 | DRASIC | SLNAC1 | TNaC1 | amiloride-sensitive cation channel 3 | ASIC3 interacts with MAGI-1B. | BIND | 15317815 |
| ACTN4 | ACTININ-4 | DKFZp686K23158 | FSGS | FSGS1 | actinin, alpha 4 | - | HPRD,BioGRID | 12042308 |
| ATN1 | B37 | D12S755E | DRPLA | NOD | atrophin 1 | Atrophin-1 interacts with AIP-3. | BIND | 9647693 |
| ATN1 | B37 | D12S755E | DRPLA | NOD | atrophin 1 | - | HPRD,BioGRID | 9647693 |
| BAI1 | FLJ41988 | brain-specific angiogenesis inhibitor 1 | - | HPRD,BioGRID | 9647739 |
| BZW1 | BZAP45 | KIAA0005 | Nbla10236 | basic leucine zipper and W2 domains 1 | Affinity Capture-MS | BioGRID | 17353931 |
| CDH1 | Arc-1 | CD324 | CDHE | ECAD | LCAM | UVO | cadherin 1, type 1, E-cadherin (epithelial) | Affinity Capture-Western | BioGRID | 10772923 |
| CTNNB1 | CTNNB | DKFZp686D02253 | FLJ25606 | FLJ37923 | catenin (cadherin-associated protein), beta 1, 88kDa | Affinity Capture-Western Reconstituted Complex Two-hybrid | BioGRID | 10772923 |
| DLL1 | DELTA1 | DL1 | Delta | delta-like 1 (Drosophila) | Delta1 interacts with MAGI-1. | BIND | 15509766 |
| FCHSD2 | KIAA0769 | NWK | SH3MD3 | FCH and double SH3 domains 2 | Affinity Capture-Western Reconstituted Complex Two-hybrid | BioGRID | 14627983 |
| IL6R | CD126 | IL-6R-1 | IL-6R-alpha | IL6RA | MGC104991 | interleukin 6 receptor | - | HPRD | 9169421 |
| LRP2 | DBS | gp330 | low density lipoprotein-related protein 2 | - | HPRD,BioGRID | 11274227 |
| NET1 | ARHGEF8 | NET1A | neuroepithelial cell transforming 1 | - | HPRD,BioGRID | 11350080 |
| RASA1 | CM-AVM | CMAVM | DKFZp434N071 | GAP | PKWS | RASA | RASGAP | p120GAP | p120RASGAP | RAS p21 protein activator (GTPase activating protein) 1 | - | HPRD | 9169421 |
| SFN | YWHAS | stratifin | Affinity Capture-MS | BioGRID | 15778465 |
| SYNPO | KIAA1029 | synaptopodin | - | HPRD,BioGRID | 12042308 |
| TP53BP2 | 53BP2 | ASPP2 | BBP | PPP1R13A | p53BP2 | tumor protein p53 binding protein, 2 | - | HPRD | 9169421 |
| WBP1 | MGC15305 | WBP-1 | WW domain binding protein 1 | Protein-peptide | BioGRID | 9169421 |
| WBP2 | MGC18269 | WBP-2 | WW domain binding protein 2 | Protein-peptide | BioGRID | 9169421 |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| KEGG TIGHT JUNCTION | 134 | 86 | All SZGR 2.0 genes in this pathway |
| RODRIGUES NTN1 TARGETS DN | 158 | 102 | All SZGR 2.0 genes in this pathway |
| WANG CLIM2 TARGETS UP | 269 | 146 | All SZGR 2.0 genes in this pathway |
| DODD NASOPHARYNGEAL CARCINOMA UP | 1821 | 933 | All SZGR 2.0 genes in this pathway |
| RODRIGUES THYROID CARCINOMA POORLY DIFFERENTIATED DN | 805 | 505 | All SZGR 2.0 genes in this pathway |
| RODRIGUES THYROID CARCINOMA ANAPLASTIC DN | 537 | 339 | All SZGR 2.0 genes in this pathway |
| GRAESSMANN APOPTOSIS BY DOXORUBICIN DN | 1781 | 1082 | All SZGR 2.0 genes in this pathway |
| KIM MYC AMPLIFICATION TARGETS UP | 201 | 127 | All SZGR 2.0 genes in this pathway |
| MATSUDA NATURAL KILLER DIFFERENTIATION | 475 | 313 | All SZGR 2.0 genes in this pathway |
| ZHAN MULTIPLE MYELOMA MS UP | 48 | 32 | All SZGR 2.0 genes in this pathway |
| LEIN ASTROCYTE MARKERS | 42 | 35 | All SZGR 2.0 genes in this pathway |
| YOSHIMURA MAPK8 TARGETS DN | 366 | 257 | All SZGR 2.0 genes in this pathway |
| BRUINS UVC RESPONSE VIA TP53 GROUP A | 898 | 516 | All SZGR 2.0 genes in this pathway |
| TORCHIA TARGETS OF EWSR1 FLI1 FUSION UP | 271 | 165 | All SZGR 2.0 genes in this pathway |
| ZWANG CLASS 1 TRANSIENTLY INDUCED BY EGF | 516 | 308 | All SZGR 2.0 genes in this pathway |
Section VI. microRNA annotation
| miRNA family | Target position | miRNA ID | miRNA seq | ||
|---|---|---|---|---|---|
| UTR start | UTR end | Match method | |||
| miR-101 | 1455 | 1462 | 1A,m8 | hsa-miR-101 | UACAGUACUGUGAUAACUGAAG |
| miR-153 | 1929 | 1935 | m8 | hsa-miR-153 | UUGCAUAGUCACAAAAGUGA |
| miR-182 | 237 | 244 | 1A,m8 | hsa-miR-182 | UUUGGCAAUGGUAGAACUCACA |
| miR-185 | 328 | 334 | m8 | hsa-miR-185brain | UGGAGAGAAAGGCAGUUC |
| miR-188 | 255 | 262 | 1A,m8 | hsa-miR-188 | CAUCCCUUGCAUGGUGGAGGGU |
| miR-205 | 576 | 583 | 1A,m8 | hsa-miR-205 | UCCUUCAUUCCACCGGAGUCUG |
| miR-23 | 1628 | 1634 | m8 | hsa-miR-23abrain | AUCACAUUGCCAGGGAUUUCC |
| hsa-miR-23bbrain | AUCACAUUGCCAGGGAUUACC | ||||
| miR-24 | 1315 | 1322 | 1A,m8 | hsa-miR-24SZ | UGGCUCAGUUCAGCAGGAACAG |
| miR-320 | 1959 | 1965 | 1A | hsa-miR-320 | AAAAGCUGGGUUGAGAGGGCGAA |
| miR-323 | 1628 | 1634 | 1A | hsa-miR-323brain | GCACAUUACACGGUCGACCUCU |
| miR-34b | 1306 | 1313 | 1A,m8 | hsa-miR-34b | UAGGCAGUGUCAUUAGCUGAUUG |
| miR-381 | 1587 | 1594 | 1A,m8 | hsa-miR-381 | UAUACAAGGGCAAGCUCUCUGU |
| miR-411 | 26 | 32 | m8 | hsa-miR-411 | AACACGGUCCACUAACCCUCAGU |
| miR-496 | 1707 | 1713 | 1A | hsa-miR-496 | AUUACAUGGCCAAUCUC |
| miR-96 | 238 | 244 | 1A | hsa-miR-96brain | UUUGGCACUAGCACAUUUUUGC |
- SZ: miRNAs which differentially expressed in brain cortex of schizophrenia patients comparing with control samples using microarray. Click here to see the list of SZ related miRNAs.
- Brain: miRNAs which are expressed in brain based on miRNA microarray expression studies. Click here to see the list of brain related miRNAs.