Gene Page: TGM5
Summary ?
| GeneID | 9333 |
| Symbol | TGM5 |
| Synonyms | PSS2|TGASE5|TGASEX|TGM6|TGMX|TGX |
| Description | transglutaminase 5 |
| Reference | MIM:603805|HGNC:HGNC:11781|Ensembl:ENSG00000104055|HPRD:11949|Vega:OTTHUMG00000176488 |
| Gene type | protein-coding |
| Map location | 15q15.2 |
| Pascal p-value | 0.003 |
| Fetal beta | -0.097 |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CV:PGCnp | Genome-wide Association Study | GWAS | |
| PMID:cooccur | High-throughput literature-search | Systematic search in PubMed for genes co-occurring with SCZ keywords. A total of 3027 genes were included. | |
| Literature | High-throughput literature-search | Co-occurance with Schizophrenia keywords: schizophrenia,schizophrenias | Click to show details |
Section I. Genetics and epigenetics annotation
 DNM table
DNM table
| Gene | Chromosome | Position | Ref | Alt | Transcript | AA change | Mutation type | Sift | CG46 | Trait | Study |
|---|---|---|---|---|---|---|---|---|---|---|---|
| TGM5 | chr15 | 44552271 | G | T | L139I | Schizophrenia | DNM:Kranz_2015 |
Section II. Transcriptome annotation
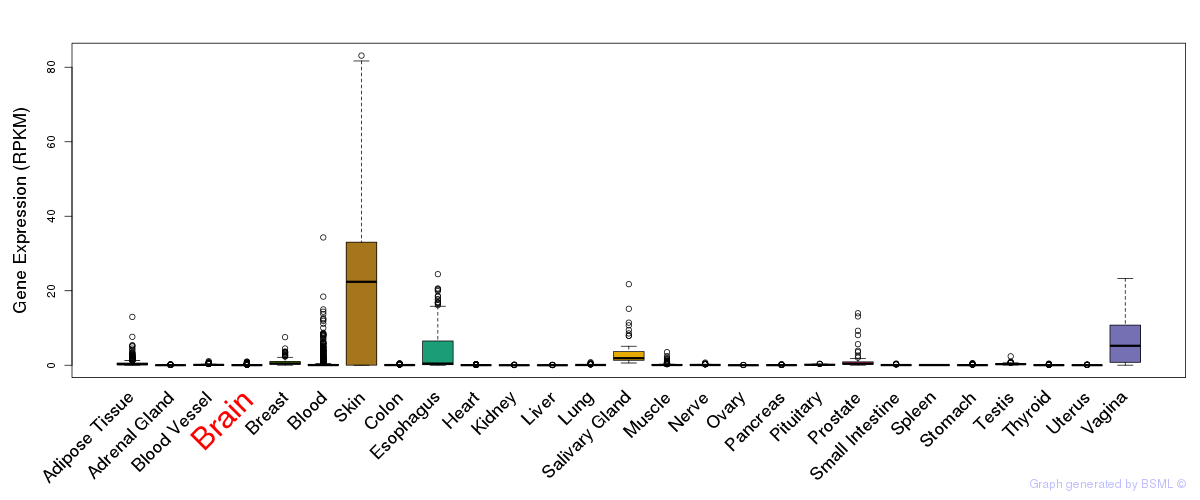
General gene expression (GTEx)
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
Top co-expressed genes in brain regions
| Top 10 positively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| SLC45A3 | 0.74 | 0.66 |
| ABCA2 | 0.73 | 0.61 |
| SLC31A2 | 0.73 | 0.64 |
| GALNTL2 | 0.72 | 0.56 |
| TTYH2 | 0.72 | 0.60 |
| PNPLA2 | 0.71 | 0.68 |
| LDB3 | 0.71 | 0.50 |
| USP54 | 0.71 | 0.62 |
| RHOG | 0.71 | 0.62 |
| SLC6A8 | 0.70 | 0.59 |
| Top 10 negatively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| SNRPG | -0.31 | -0.37 |
| BTBD17 | -0.31 | -0.29 |
| FAM36A | -0.30 | -0.26 |
| SPINK8 | -0.29 | -0.29 |
| SPDYA | -0.29 | -0.29 |
| FAM159B | -0.29 | -0.38 |
| C9orf46 | -0.29 | -0.29 |
| AC087071.1 | -0.28 | -0.24 |
| RP11-3P17.1 | -0.27 | -0.31 |
| RP9P | -0.27 | -0.30 |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| TAKEDA TARGETS OF NUP98 HOXA9 FUSION 8D UP | 157 | 91 | All SZGR 2.0 genes in this pathway |
| TAKEDA TARGETS OF NUP98 HOXA9 FUSION 10D UP | 194 | 122 | All SZGR 2.0 genes in this pathway |
| TAKEDA TARGETS OF NUP98 HOXA9 FUSION 16D UP | 175 | 108 | All SZGR 2.0 genes in this pathway |
| JAZAG TGFB1 SIGNALING UP | 108 | 69 | All SZGR 2.0 genes in this pathway |
| PUJANA BRCA1 PCC NETWORK | 1652 | 1023 | All SZGR 2.0 genes in this pathway |
| PUJANA ATM PCC NETWORK | 1442 | 892 | All SZGR 2.0 genes in this pathway |
| IVANOVA HEMATOPOIESIS STEM CELL LONG TERM | 302 | 191 | All SZGR 2.0 genes in this pathway |
| VALK AML CLUSTER 16 | 26 | 17 | All SZGR 2.0 genes in this pathway |
| VALK AML WITH 11Q23 REARRANGED | 22 | 13 | All SZGR 2.0 genes in this pathway |
| YAGI AML WITH T 9 11 TRANSLOCATION | 130 | 87 | All SZGR 2.0 genes in this pathway |
| YAGI AML WITH 11Q23 REARRANGED | 351 | 238 | All SZGR 2.0 genes in this pathway |
| MIKKELSEN NPC LCP WITH H3K4ME3 | 58 | 34 | All SZGR 2.0 genes in this pathway |
| NABA ECM REGULATORS | 238 | 125 | All SZGR 2.0 genes in this pathway |
| NABA MATRISOME ASSOCIATED | 753 | 411 | All SZGR 2.0 genes in this pathway |
| NABA MATRISOME | 1028 | 559 | All SZGR 2.0 genes in this pathway |