Gene Page: CHST10
Summary ?
| GeneID | 9486 |
| Symbol | CHST10 |
| Synonyms | HNK-1ST|HNK1ST |
| Description | carbohydrate sulfotransferase 10 |
| Reference | MIM:606376|HGNC:HGNC:19650|Ensembl:ENSG00000115526|HPRD:07332|Vega:OTTHUMG00000130669 |
| Gene type | protein-coding |
| Map location | 2q11.2 |
| Pascal p-value | 0.364 |
| Sherlock p-value | 0.509 |
| Fetal beta | 1.073 |
| eGene | Cerebellar Hemisphere Cerebellum |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CV:PGCnp | Genome-wide Association Study | GWAS | |
| PMID:cooccur | High-throughput literature-search | Systematic search in PubMed for genes co-occurring with SCZ keywords. A total of 3027 genes were included. | |
| GSMA_I | Genome scan meta-analysis | Psr: 0.0004 | |
| Literature | High-throughput literature-search | Co-occurance with Schizophrenia keywords: schizophrenia,schizophrenias | Click to show details |
Section I. Genetics and epigenetics annotation
Section II. Transcriptome annotation
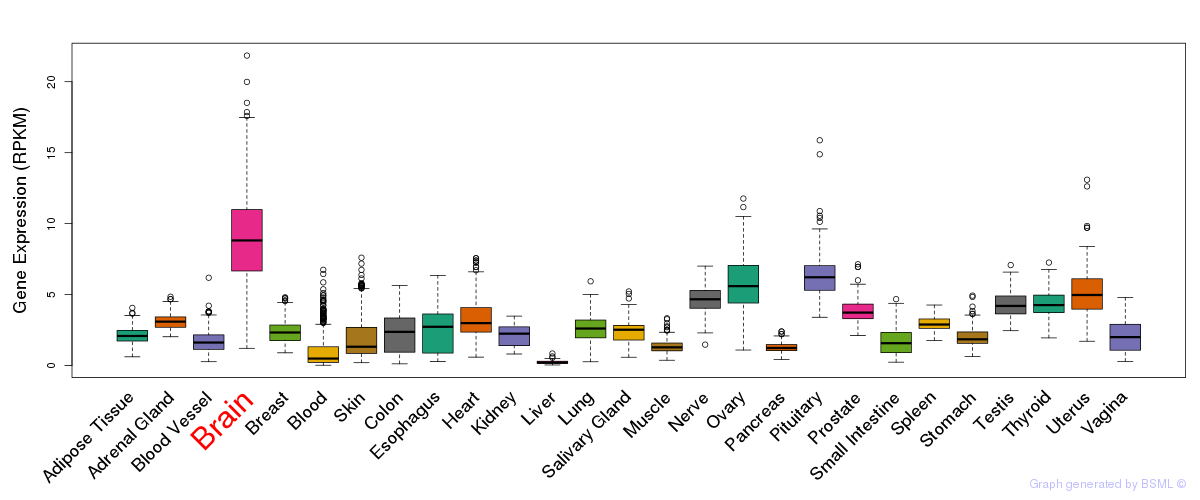
General gene expression (GTEx)
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
Top co-expressed genes in brain regions
| Top 10 positively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| GRAMD3 | 0.81 | 0.82 |
| ALDH6A1 | 0.78 | 0.75 |
| TPP1 | 0.77 | 0.76 |
| TMBIM1 | 0.76 | 0.78 |
| DTNA | 0.76 | 0.76 |
| SPARCL1 | 0.75 | 0.78 |
| MRO | 0.74 | 0.75 |
| ANXA4 | 0.74 | 0.76 |
| GALNTL2 | 0.74 | 0.78 |
| KCNJ10 | 0.73 | 0.78 |
| Top 10 negatively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| NKIRAS2 | -0.55 | -0.54 |
| HN1 | -0.53 | -0.57 |
| KIAA1949 | -0.52 | -0.51 |
| TUBB | -0.52 | -0.54 |
| NR2C2AP | -0.52 | -0.60 |
| ZNF821 | -0.52 | -0.53 |
| MED19 | -0.52 | -0.61 |
| PAFAH1B3 | -0.52 | -0.62 |
| TUBB2B | -0.52 | -0.51 |
| MPP3 | -0.52 | -0.51 |
Section III. Gene Ontology annotation
| Molecular function | GO term | Evidence | Neuro keywords | PubMed ID |
|---|---|---|---|---|
| GO:0016740 | transferase activity | IEA | - | |
| GO:0008146 | sulfotransferase activity | TAS | 9478973 | |
| Biological process | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0007155 | cell adhesion | TAS | 9478973 | |
| GO:0005975 | carbohydrate metabolic process | IEA | - | |
| Cellular component | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0000139 | Golgi membrane | IEA | - | |
| GO:0005794 | Golgi apparatus | TAS | 9478973 | |
| GO:0005624 | membrane fraction | TAS | 9478973 | |
| GO:0016020 | membrane | IEA | - | |
| GO:0016021 | integral to membrane | IEA | - |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| GRAESSMANN APOPTOSIS BY DOXORUBICIN DN | 1781 | 1082 | All SZGR 2.0 genes in this pathway |
| RICKMAN METASTASIS UP | 344 | 180 | All SZGR 2.0 genes in this pathway |
| BROWNE HCMV INFECTION 20HR UP | 240 | 152 | All SZGR 2.0 genes in this pathway |
| MAHAJAN RESPONSE TO IL1A DN | 76 | 57 | All SZGR 2.0 genes in this pathway |
| YOSHIMURA MAPK8 TARGETS UP | 1305 | 895 | All SZGR 2.0 genes in this pathway |
| KIM ALL DISORDERS CALB1 CORR UP | 548 | 370 | All SZGR 2.0 genes in this pathway |
| CHYLA CBFA2T3 TARGETS DN | 242 | 146 | All SZGR 2.0 genes in this pathway |
| KASLER HDAC7 TARGETS 1 UP | 194 | 133 | All SZGR 2.0 genes in this pathway |
Section VI. microRNA annotation
| miRNA family | Target position | miRNA ID | miRNA seq | ||
|---|---|---|---|---|---|
| UTR start | UTR end | Match method | |||
| miR-137 | 1092 | 1098 | 1A | hsa-miR-137 | UAUUGCUUAAGAAUACGCGUAG |
| miR-182 | 1374 | 1380 | 1A | hsa-miR-182 | UUUGGCAAUGGUAGAACUCACA |
| miR-23 | 1348 | 1355 | 1A,m8 | hsa-miR-23abrain | AUCACAUUGCCAGGGAUUUCC |
| hsa-miR-23bbrain | AUCACAUUGCCAGGGAUUACC | ||||
| miR-323 | 1348 | 1354 | 1A | hsa-miR-323brain | GCACAUUACACGGUCGACCUCU |
| miR-96 | 1373 | 1380 | 1A,m8 | hsa-miR-96brain | UUUGGCACUAGCACAUUUUUGC |
- SZ: miRNAs which differentially expressed in brain cortex of schizophrenia patients comparing with control samples using microarray. Click here to see the list of SZ related miRNAs.
- Brain: miRNAs which are expressed in brain based on miRNA microarray expression studies. Click here to see the list of brain related miRNAs.