Gene Page: ITIH4
Summary ?
| GeneID | 3700 |
| Symbol | ITIH4 |
| Synonyms | GP120|H4P|IHRP|ITI-HC4|ITIHL1|PK-120|PK120 |
| Description | inter-alpha-trypsin inhibitor heavy chain family member 4 |
| Reference | MIM:600564|HGNC:HGNC:6169|Ensembl:ENSG00000055955|HPRD:02781|Vega:OTTHUMG00000159023 |
| Gene type | protein-coding |
| Map location | 3p21.1 |
| Pascal p-value | 1.521E-10 |
| Sherlock p-value | 0.118 |
| Fetal beta | -0.564 |
| eGene | Myers' cis & trans Meta |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CV:GWAScat | Genome-wide Association Studies | This data set includes 560 SNPs associated with schizophrenia. A total of 486 genes were mapped to these SNPs within 50kb. | |
| CV:GWASdb | Genome-wide Association Studies | GWASdb records for schizophrenia | |
| CV:PGC128 | Genome-wide Association Study | A multi-stage schizophrenia GWAS of up to 36,989 cases and 113,075 controls. Reported by the Schizophrenia Working Group of PGC. 128 independent associations spanning 108 loci | |
| CV:PGCnp | Genome-wide Association Study | GWAS | |
| LK:YES | Genome-wide Association Study | This data set included 99 genes mapped to the 22 regions. The 24 leading SNPs were also included in CV:Ripke_2013 | |
| PMID:cooccur | High-throughput literature-search | Systematic search in PubMed for genes co-occurring with SCZ keywords. A total of 3027 genes were included. | |
| Literature | High-throughput literature-search | Co-occurance with Schizophrenia keywords: schizophrenia,schizophrenic,schizophrenias | Click to show details |
Section I. Genetics and epigenetics annotation
 CV:PGC128
CV:PGC128
| SNP ID | Chromosome | Position | Allele | P | Function | Gene | Up/Down Distance |
|---|---|---|---|---|---|---|---|
| rs2535627 | chr3 | 52845105 | TC | 3.956E-11 | intergenic | ITIH3,ITIH4 | dist=2080;dist=1901 |
 eQTL annotation
eQTL annotation
| SNP ID | Chromosome | Position | eGene | Gene Entrez ID | pvalue | qvalue | TSS distance | eQTL type |
|---|---|---|---|---|---|---|---|---|
| rs1961958 | chr3 | 52585989 | ITIH4 | 3700 | 0.18 | cis | ||
| rs6778844 | chr3 | 52596397 | ITIH4 | 3700 | 0.02 | cis | ||
| rs2336545 | chr3 | 52787602 | ITIH4 | 3700 | 0.18 | cis | ||
| rs2240920 | chr3 | 52831008 | ITIH4 | 3700 | 0.1 | cis | ||
| rs2276817 | chr3 | 52860935 | ITIH4 | 3700 | 0 | cis | ||
| rs3733047 | chr3 | 52871928 | ITIH4 | 3700 | 0.03 | cis | ||
| rs6803519 | chr3 | 52889770 | ITIH4 | 3700 | 0 | cis | ||
| rs2581815 | chr3 | 52982158 | ITIH4 | 3700 | 0.01 | cis | ||
| rs2336669 | chr3 | 52994137 | ITIH4 | 3700 | 0.01 | cis | ||
| rs4519686 | chr3 | 52997870 | ITIH4 | 3700 | 0.04 | cis | ||
| rs17304995 | chr3 | 53070754 | ITIH4 | 3700 | 0.01 | cis | ||
| rs2276817 | chr3 | 52860935 | ITIH4 | 3700 | 0.11 | trans | ||
| rs6803519 | chr3 | 52889770 | ITIH4 | 3700 | 0.19 | trans |
Section II. Transcriptome annotation
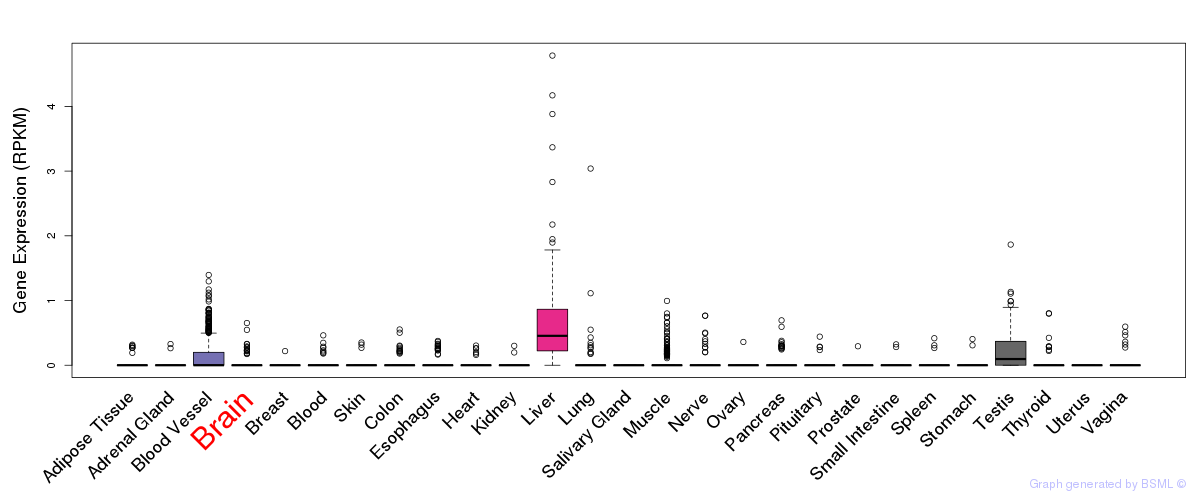
General gene expression (GTEx)
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
Top co-expressed genes in brain regions
| Top 10 positively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| DDIT4 | 0.69 | 0.45 |
| KDM3A | 0.66 | 0.58 |
| TMPO | 0.66 | 0.55 |
| HIST2H2BE | 0.65 | 0.56 |
| ZFP36L1 | 0.65 | 0.57 |
| HES1 | 0.64 | 0.43 |
| MCL1 | 0.64 | 0.61 |
| VEGFA | 0.64 | 0.53 |
| ZNF143 | 0.63 | 0.52 |
| SORD | 0.63 | 0.56 |
| Top 10 negatively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| CLDND2 | -0.40 | -0.47 |
| C5orf53 | -0.40 | -0.46 |
| AF347015.21 | -0.39 | -0.49 |
| IFI27 | -0.39 | -0.44 |
| MT-CO2 | -0.37 | -0.45 |
| AF347015.27 | -0.36 | -0.44 |
| AF347015.31 | -0.36 | -0.42 |
| C1orf54 | -0.36 | -0.44 |
| PSMB9 | -0.35 | -0.37 |
| HIGD1B | -0.35 | -0.41 |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| CORRE MULTIPLE MYELOMA UP | 74 | 45 | All SZGR 2.0 genes in this pathway |
| HOEBEKE LYMPHOID STEM CELL UP | 95 | 64 | All SZGR 2.0 genes in this pathway |
| TARTE PLASMA CELL VS PLASMABLAST UP | 398 | 262 | All SZGR 2.0 genes in this pathway |
| HSIAO LIVER SPECIFIC GENES | 244 | 154 | All SZGR 2.0 genes in this pathway |
| IGLESIAS E2F TARGETS DN | 16 | 11 | All SZGR 2.0 genes in this pathway |
| WANG CISPLATIN RESPONSE AND XPC DN | 228 | 146 | All SZGR 2.0 genes in this pathway |
| BLALOCK ALZHEIMERS DISEASE UP | 1691 | 1088 | All SZGR 2.0 genes in this pathway |
| LEE LIVER CANCER | 49 | 29 | All SZGR 2.0 genes in this pathway |
| STEARMAN TUMOR FIELD EFFECT UP | 36 | 22 | All SZGR 2.0 genes in this pathway |
| STEARMAN LUNG CANCER EARLY VS LATE UP | 125 | 89 | All SZGR 2.0 genes in this pathway |
| SWEET LUNG CANCER KRAS UP | 491 | 316 | All SZGR 2.0 genes in this pathway |
| BOYAULT LIVER CANCER SUBCLASS G123 DN | 51 | 30 | All SZGR 2.0 genes in this pathway |
| HOSHIDA LIVER CANCER SUBCLASS S3 | 266 | 180 | All SZGR 2.0 genes in this pathway |
| CAIRO HEPATOBLASTOMA DN | 267 | 160 | All SZGR 2.0 genes in this pathway |
| CAIRO HEPATOBLASTOMA CLASSES DN | 210 | 141 | All SZGR 2.0 genes in this pathway |
| CAIRO LIVER DEVELOPMENT DN | 222 | 141 | All SZGR 2.0 genes in this pathway |
| FEVR CTNNB1 TARGETS UP | 682 | 433 | All SZGR 2.0 genes in this pathway |
| WAKABAYASHI ADIPOGENESIS PPARG BOUND 8D | 658 | 397 | All SZGR 2.0 genes in this pathway |
| NABA ECM REGULATORS | 238 | 125 | All SZGR 2.0 genes in this pathway |
| NABA MATRISOME ASSOCIATED | 753 | 411 | All SZGR 2.0 genes in this pathway |
| NABA MATRISOME | 1028 | 559 | All SZGR 2.0 genes in this pathway |