Gene Page: SAE1
Summary ?
| GeneID | 10055 |
| Symbol | SAE1 |
| Synonyms | AOS1|HSPC140|SUA1|UBLE1A |
| Description | SUMO1 activating enzyme subunit 1 |
| Reference | MIM:613294|HGNC:HGNC:30660|Ensembl:ENSG00000142230|HPRD:18010|Vega:OTTHUMG00000183464 |
| Gene type | protein-coding |
| Map location | 19q13.32 |
| Pascal p-value | 0.11 |
| Sherlock p-value | 0.595 |
| Fetal beta | 0.747 |
| DMG | 1 (# studies) |
| Support | G2Cdb.humanPSD G2Cdb.humanPSP |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CV:PGCnp | Genome-wide Association Study | GWAS | |
| DMG:Wockner_2014 | Genome-wide DNA methylation analysis | This dataset includes 4641 differentially methylated probes corresponding to 2929 unique genes between schizophrenia patients (n=24) and controls (n=24). | 1 |
Section I. Genetics and epigenetics annotation
 Differentially methylated gene
Differentially methylated gene
| Probe | Chromosome | Position | Nearest gene | P (dis) | Beta (dis) | FDR (dis) | Study |
|---|---|---|---|---|---|---|---|
| cg16041550 | 19 | 47633739 | SAE1 | 3.318E-4 | 0.376 | 0.041 | DMG:Wockner_2014 |
Section II. Transcriptome annotation
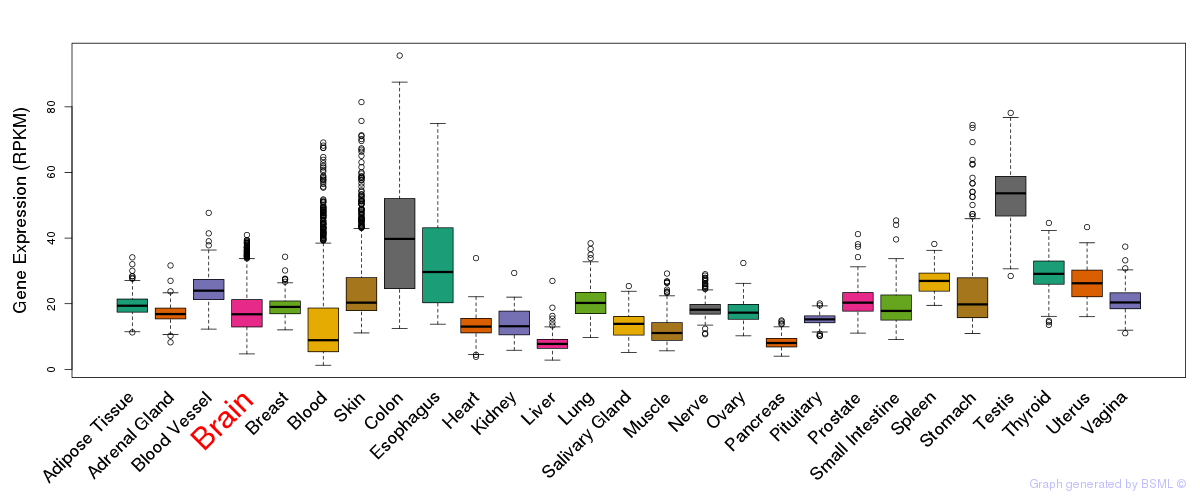
General gene expression (GTEx)
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
Top co-expressed genes in brain regions
| Top 10 positively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| ZXDC | 0.91 | 0.87 |
| CD276 | 0.91 | 0.87 |
| ABL1 | 0.90 | 0.85 |
| CCNJ | 0.90 | 0.85 |
| MOBKL2A | 0.90 | 0.87 |
| EPHB2 | 0.90 | 0.85 |
| DLG5 | 0.90 | 0.83 |
| EHMT1 | 0.90 | 0.85 |
| ZNF319 | 0.89 | 0.86 |
| ZNF532 | 0.89 | 0.86 |
| Top 10 negatively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| AF347015.27 | -0.68 | -0.81 |
| AF347015.31 | -0.67 | -0.81 |
| MT-CO2 | -0.66 | -0.81 |
| C5orf53 | -0.65 | -0.69 |
| AF347015.8 | -0.64 | -0.80 |
| MT-CYB | -0.64 | -0.79 |
| AF347015.33 | -0.64 | -0.77 |
| AF347015.15 | -0.62 | -0.78 |
| HLA-F | -0.61 | -0.62 |
| AIFM3 | -0.60 | -0.62 |
Section IV. Protein-protein interaction annotation
| Interactors | Aliases B | Official full name B | Experimental | Source | PubMed ID |
|---|---|---|---|---|---|
| CYP1B1 | CP1B | GLC3A | P4501B1 | cytochrome P450, family 1, subfamily B, polypeptide 1 | Affinity Capture-MS | BioGRID | 17353931 |
| FKBP4 | FKBP52 | FKBP59 | HBI | Hsp56 | PPIase | p52 | FK506 binding protein 4, 59kDa | Affinity Capture-MS | BioGRID | 17353931 |
| KIAA1409 | FLJ43337 | KIAA1409 | Affinity Capture-MS | BioGRID | 17353931 |
| RANBP2 | NUP358 | TRP1 | TRP2 | RAN binding protein 2 | Biochemical Activity | BioGRID | 11792325 |
| RANGAP1 | Fug1 | KIAA1835 | MGC20266 | SD | Ran GTPase activating protein 1 | - | HPRD | 9920803 |
| SP100 | DKFZp686E07254 | FLJ00340 | FLJ34579 | SP100 nuclear antigen | Biochemical Activity | BioGRID | 11792325 |
| SUMO1 | DAP-1 | GMP1 | OFC10 | PIC1 | SENP2 | SMT3 | SMT3C | SMT3H3 | SUMO-1 | UBL1 | SMT3 suppressor of mif two 3 homolog 1 (S. cerevisiae) | Reconstituted Complex | BioGRID | 12924945 |
| SUMO1 | DAP-1 | GMP1 | OFC10 | PIC1 | SENP2 | SMT3 | SMT3C | SMT3H3 | SUMO-1 | UBL1 | SMT3 suppressor of mif two 3 homolog 1 (S. cerevisiae) | - | HPRD | 10187858 |
| SUMO3 | SMT3A | SMT3H1 | SUMO-3 | SMT3 suppressor of mif two 3 homolog 3 (S. cerevisiae) | Affinity Capture-MS | BioGRID | 17353931 |
| SUMO3 | SMT3A | SMT3H1 | SUMO-3 | SMT3 suppressor of mif two 3 homolog 3 (S. cerevisiae) | - | HPRD | 12924945 |
| TP53 | FLJ92943 | LFS1 | TRP53 | p53 | tumor protein p53 | - | HPRD | 15327968 |
| UBA2 | ARX | FLJ13058 | HRIHFB2115 | SAE2 | ubiquitin-like modifier activating enzyme 2 | Sua1 interacts with hUba2. | BIND | 9920803 |
| UBA2 | ARX | FLJ13058 | HRIHFB2115 | SAE2 | ubiquitin-like modifier activating enzyme 2 | Affinity Capture-MS Biochemical Activity in vitro Two-hybrid | BioGRID | 10187858 |10217437 |16189514 |17353931 |
| UBA2 | ARX | FLJ13058 | HRIHFB2115 | SAE2 | ubiquitin-like modifier activating enzyme 2 | - | HPRD | 9920803 |10187858 |10217437 |
| UBA2 | ARX | FLJ13058 | HRIHFB2115 | SAE2 | ubiquitin-like modifier activating enzyme 2 | SAE1 interacts with SAE2. This interaction was modeled on a demonstrated interaction between SAE1 from an unspecified species and SAE2 from an unspecified species. | BIND | 15546615 |
| UBE2E3 | UBCH9 | UbcM2 | ubiquitin-conjugating enzyme E2E 3 (UBC4/5 homolog, yeast) | - | HPRD | 10187858 |
| UBE2I | C358B7.1 | P18 | UBC9 | ubiquitin-conjugating enzyme E2I (UBC9 homolog, yeast) | Affinity Capture-MS | BioGRID | 17353931 |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| KEGG UBIQUITIN MEDIATED PROTEOLYSIS | 138 | 98 | All SZGR 2.0 genes in this pathway |
| REACTOME IMMUNE SYSTEM | 933 | 616 | All SZGR 2.0 genes in this pathway |
| REACTOME ADAPTIVE IMMUNE SYSTEM | 539 | 350 | All SZGR 2.0 genes in this pathway |
| REACTOME CLASS I MHC MEDIATED ANTIGEN PROCESSING PRESENTATION | 251 | 156 | All SZGR 2.0 genes in this pathway |
| REACTOME ANTIGEN PROCESSING UBIQUITINATION PROTEASOME DEGRADATION | 212 | 129 | All SZGR 2.0 genes in this pathway |
| CHEMNITZ RESPONSE TO PROSTAGLANDIN E2 UP | 146 | 86 | All SZGR 2.0 genes in this pathway |
| BASAKI YBX1 TARGETS UP | 290 | 177 | All SZGR 2.0 genes in this pathway |
| OSMAN BLADDER CANCER UP | 404 | 246 | All SZGR 2.0 genes in this pathway |
| KINSEY TARGETS OF EWSR1 FLII FUSION UP | 1278 | 748 | All SZGR 2.0 genes in this pathway |
| LINDGREN BLADDER CANCER CLUSTER 1 DN | 378 | 231 | All SZGR 2.0 genes in this pathway |
| LINDGREN BLADDER CANCER CLUSTER 3 UP | 329 | 196 | All SZGR 2.0 genes in this pathway |
| MARKEY RB1 ACUTE LOF DN | 228 | 137 | All SZGR 2.0 genes in this pathway |
| GRAESSMANN APOPTOSIS BY DOXORUBICIN DN | 1781 | 1082 | All SZGR 2.0 genes in this pathway |
| BERENJENO TRANSFORMED BY RHOA UP | 536 | 340 | All SZGR 2.0 genes in this pathway |
| NUYTTEN EZH2 TARGETS DN | 1024 | 594 | All SZGR 2.0 genes in this pathway |
| BENPORATH MYC MAX TARGETS | 775 | 494 | All SZGR 2.0 genes in this pathway |
| ZUCCHI METASTASIS UP | 43 | 24 | All SZGR 2.0 genes in this pathway |
| IVANOVA HEMATOPOIESIS EARLY PROGENITOR | 532 | 309 | All SZGR 2.0 genes in this pathway |
| BLALOCK ALZHEIMERS DISEASE UP | 1691 | 1088 | All SZGR 2.0 genes in this pathway |
| GERHOLD ADIPOGENESIS DN | 64 | 44 | All SZGR 2.0 genes in this pathway |
| MARSON BOUND BY E2F4 UNSTIMULATED | 728 | 415 | All SZGR 2.0 genes in this pathway |
| RIZKI TUMOR INVASIVENESS 3D UP | 210 | 124 | All SZGR 2.0 genes in this pathway |
| FOURNIER ACINAR DEVELOPMENT LATE 2 | 277 | 172 | All SZGR 2.0 genes in this pathway |
| CAIRO HEPATOBLASTOMA CLASSES UP | 605 | 377 | All SZGR 2.0 genes in this pathway |
| DANG BOUND BY MYC | 1103 | 714 | All SZGR 2.0 genes in this pathway |
| YAO TEMPORAL RESPONSE TO PROGESTERONE CLUSTER 17 | 181 | 101 | All SZGR 2.0 genes in this pathway |
| PILON KLF1 TARGETS DN | 1972 | 1213 | All SZGR 2.0 genes in this pathway |
| GOBERT OLIGODENDROCYTE DIFFERENTIATION DN | 1080 | 713 | All SZGR 2.0 genes in this pathway |