Gene Page: AKAP9
Summary ?
| GeneID | 10142 |
| Symbol | AKAP9 |
| Synonyms | AKAP-9|AKAP350|AKAP450|CG-NAP|HYPERION|LQT11|MU-RMS-40.16A|PPP1R45|PRKA9|YOTIAO |
| Description | A-kinase anchoring protein 9 |
| Reference | MIM:604001|HGNC:HGNC:379|Ensembl:ENSG00000127914|HPRD:04921|Vega:OTTHUMG00000131127 |
| Gene type | protein-coding |
| Map location | 7q21-q22 |
| Pascal p-value | 0.111 |
| Fetal beta | 1.52 |
| Support | G2Cdb.humanNRC CompositeSet Darnell FMRP targets Potential synaptic genes |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CV:GWASdb | Genome-wide Association Studies | GWASdb records for schizophrenia | |
| CV:PGCnp | Genome-wide Association Study | GWAS | |
| DNM:Fromer_2014 | Whole Exome Sequencing analysis | This study reported a WES study of 623 schizophrenia trios, reporting DNMs using genomic DNA. | |
| PMID:cooccur | High-throughput literature-search | Systematic search in PubMed for genes co-occurring with SCZ keywords. A total of 3027 genes were included. | |
| Literature | High-throughput literature-search | Co-occurance with Schizophrenia keywords: schizophrenia,schizophrenic,schizophrenias | Click to show details |
| GO_Annotation | Mapping neuro-related keywords to Gene Ontology annotations | Hits with neuro-related keywords: 2 | |
| Network | Shortest path distance of core genes in the Human protein-protein interaction network | Contribution to shortest path in PPI network: 0.3189 |
Section I. Genetics and epigenetics annotation
 DNM table
DNM table
| Gene | Chromosome | Position | Ref | Alt | Transcript | AA change | Mutation type | Sift | CG46 | Trait | Study |
|---|---|---|---|---|---|---|---|---|---|---|---|
| AKAP9 | chr7 | 91674467 | A | T | NM_005751 NM_147185 | p.1770I>F p.1770I>F | missense missense | Schizophrenia | DNM:Fromer_2014 |
Section II. Transcriptome annotation
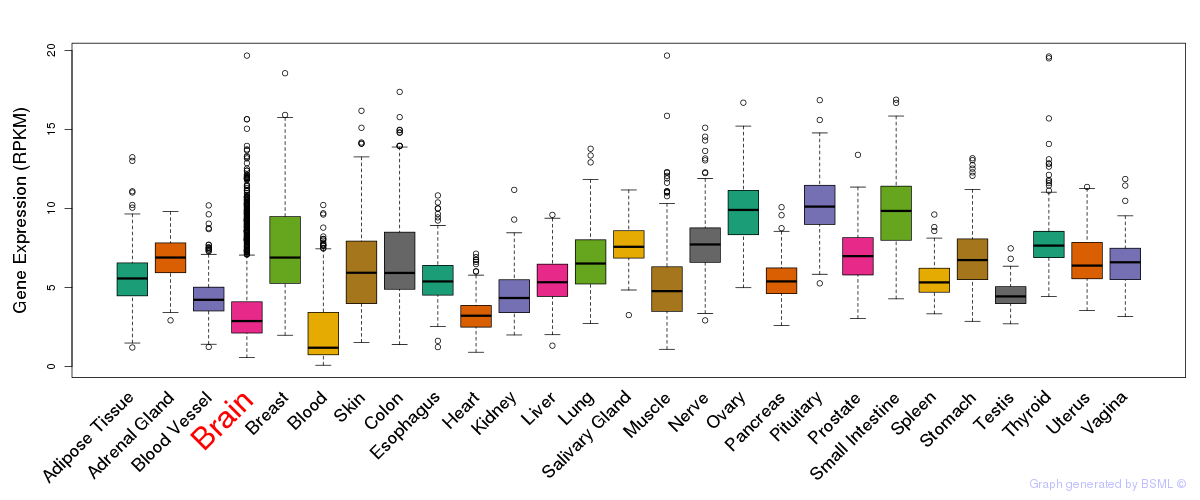
General gene expression (GTEx)
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
Top co-expressed genes in brain regions
| Top 10 positively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| RHBDF2 | 0.47 | 0.52 |
| HMOX1 | 0.46 | 0.51 |
| C10orf54 | 0.46 | 0.51 |
| SERPINA1 | 0.45 | 0.40 |
| CD40 | 0.45 | 0.49 |
| TMBIM1 | 0.45 | 0.48 |
| CASS4 | 0.45 | 0.47 |
| SPI1 | 0.45 | 0.47 |
| NLRC5 | 0.44 | 0.46 |
| RAC2 | 0.44 | 0.47 |
| Top 10 negatively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| RTF1 | -0.30 | -0.31 |
| RP9 | -0.30 | -0.34 |
| MPHOSPH10 | -0.29 | -0.30 |
| CIR1 | -0.29 | -0.32 |
| RBMX2 | -0.29 | -0.33 |
| NOVA1 | -0.29 | -0.29 |
| THOC2 | -0.29 | -0.31 |
| RIOK1 | -0.28 | -0.29 |
| RSBN1L | -0.28 | -0.30 |
| ZNF30 | -0.28 | -0.30 |
Section III. Gene Ontology annotation
| Molecular function | GO term | Evidence | Neuro keywords | PubMed ID |
|---|---|---|---|---|
| GO:0005102 | receptor binding | TAS | Neurotransmitter (GO term level: 4) | 9482789 |
| GO:0016301 | kinase activity | IEA | - | |
| Biological process | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0007268 | synaptic transmission | TAS | neuron, Synap, Neurotransmitter (GO term level: 6) | 9482789 |
| GO:0007165 | signal transduction | TAS | 10390370 | |
| GO:0006810 | transport | TAS | 10390370 | |
| Cellular component | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0005794 | Golgi apparatus | IEA | - | |
| GO:0005813 | centrosome | TAS | 10202149 | |
| GO:0005856 | cytoskeleton | TAS | 9482789 | |
| GO:0005737 | cytoplasm | IEA | - |
Section IV. Protein-protein interaction annotation
| Interactors | Aliases B | Official full name B | Experimental | Source | PubMed ID |
|---|---|---|---|---|---|
| CALM1 | CALML2 | CAMI | DD132 | PHKD | calmodulin 1 (phosphorylase kinase, delta) | Affinity Capture-Western Reconstituted Complex | BioGRID | 12221128 |
| CALM2 | CAMII | PHKD | PHKD2 | calmodulin 2 (phosphorylase kinase, delta) | Two-hybrid | BioGRID | 12221128 |
| CLIC1 | G6 | NCC27 | chloride intracellular channel 1 | - | HPRD,BioGRID | 12163479 |
| CSNK1D | HCKID | casein kinase 1, delta | - | HPRD,BioGRID | 12270714 |
| CSNK1E | HCKIE | MGC10398 | casein kinase 1, epsilon | - | HPRD | 12270714 |
| FNBP1 | FBP17 | KIAA0554 | MGC126804 | formin binding protein 1 | - | HPRD,BioGRID | 15047863 |
| GRIN1 | NMDA1 | NMDAR1 | NR1 | glutamate receptor, ionotropic, N-methyl D-aspartate 1 | - | HPRD | 9482789|10862698 |
| GRIN1 | NMDA1 | NMDAR1 | NR1 | glutamate receptor, ionotropic, N-methyl D-aspartate 1 | - | HPRD | 10862698 |
| KCNQ1 | ATFB1 | FLJ26167 | JLNS1 | KCNA8 | KCNA9 | KVLQT1 | Kv1.9 | Kv7.1 | LQT | LQT1 | RWS | SQT2 | WRS | potassium voltage-gated channel, KQT-like subfamily, member 1 | - | HPRD,BioGRID | 11799244 |
| PCNT | KEN | MOPD2 | PCN | PCNT2 | PCNTB | PCTN2 | SCKL4 | pericentrin | - | HPRD | 12221128 |
| PKN1 | DBK | MGC46204 | PAK1 | PKN | PKN-ALPHA | PRK1 | PRKCL1 | protein kinase N1 | - | HPRD,BioGRID | 10358086 |
| PPP1CA | MGC15877 | MGC1674 | PP-1A | PPP1A | protein phosphatase 1, catalytic subunit, alpha isoform | - | HPRD | 10358086 |
| PPP2R1B | MGC26454 | PR65B | protein phosphatase 2 (formerly 2A), regulatory subunit A, beta isoform | - | HPRD | 10358086 |
| PRKAR2A | MGC3606 | PKR2 | PRKAR2 | protein kinase, cAMP-dependent, regulatory, type II, alpha | - | HPRD,BioGRID | 10358086 |
| PRKAR2A | MGC3606 | PKR2 | PRKAR2 | protein kinase, cAMP-dependent, regulatory, type II, alpha | - | HPRD | 10358086 |11799244 |
| PRKAR2B | PRKAR2 | RII-BETA | protein kinase, cAMP-dependent, regulatory, type II, beta | - | HPRD | 11799244 |
| PRKCE | MGC125656 | MGC125657 | PKCE | nPKC-epsilon | protein kinase C, epsilon | - | HPRD | 10945988 |
| TACC3 | ERIC1 | MGC117382 | MGC133242 | transforming, acidic coiled-coil containing protein 3 | - | HPRD,BioGRID | 12015314 |
| TRIP10 | CIP4 | HSTP | STOT | STP | thyroid hormone receptor interactor 10 | Reconstituted Complex Two-hybrid | BioGRID | 15047863 |
| TSNAX | TRAX | translin-associated factor X | - | HPRD | 12036294 |
| TUBGCP3 | GCP3 | SPBC98 | Spc98p | tubulin, gamma complex associated protein 3 | - | HPRD,BioGRID | 12221128 |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| BIOCARTA AKAPCENTROSOME PATHWAY | 15 | 14 | All SZGR 2.0 genes in this pathway |
| REACTOME CELL CYCLE | 421 | 253 | All SZGR 2.0 genes in this pathway |
| REACTOME TRANSMISSION ACROSS CHEMICAL SYNAPSES | 186 | 155 | All SZGR 2.0 genes in this pathway |
| REACTOME NEURONAL SYSTEM | 279 | 221 | All SZGR 2.0 genes in this pathway |
| REACTOME CELL CYCLE MITOTIC | 325 | 185 | All SZGR 2.0 genes in this pathway |
| REACTOME RECRUITMENT OF MITOTIC CENTROSOME PROTEINS AND COMPLEXES | 66 | 43 | All SZGR 2.0 genes in this pathway |
| REACTOME LOSS OF NLP FROM MITOTIC CENTROSOMES | 59 | 38 | All SZGR 2.0 genes in this pathway |
| REACTOME NEUROTRANSMITTER RECEPTOR BINDING AND DOWNSTREAM TRANSMISSION IN THE POSTSYNAPTIC CELL | 137 | 110 | All SZGR 2.0 genes in this pathway |
| REACTOME RAS ACTIVATION UOPN CA2 INFUX THROUGH NMDA RECEPTOR | 17 | 15 | All SZGR 2.0 genes in this pathway |
| REACTOME ACTIVATION OF NMDA RECEPTOR UPON GLUTAMATE BINDING AND POSTSYNAPTIC EVENTS | 37 | 32 | All SZGR 2.0 genes in this pathway |
| REACTOME CREB PHOSPHORYLATION THROUGH THE ACTIVATION OF RAS | 27 | 23 | All SZGR 2.0 genes in this pathway |
| REACTOME POST NMDA RECEPTOR ACTIVATION EVENTS | 33 | 28 | All SZGR 2.0 genes in this pathway |
| REACTOME UNBLOCKING OF NMDA RECEPTOR GLUTAMATE BINDING AND ACTIVATION | 15 | 14 | All SZGR 2.0 genes in this pathway |
| REACTOME CREB PHOSPHORYLATION THROUGH THE ACTIVATION OF CAMKII | 15 | 14 | All SZGR 2.0 genes in this pathway |
| REACTOME MITOTIC G2 G2 M PHASES | 81 | 50 | All SZGR 2.0 genes in this pathway |
| ONKEN UVEAL MELANOMA UP | 783 | 507 | All SZGR 2.0 genes in this pathway |
| WILCOX RESPONSE TO PROGESTERONE DN | 66 | 44 | All SZGR 2.0 genes in this pathway |
| DOANE RESPONSE TO ANDROGEN DN | 241 | 146 | All SZGR 2.0 genes in this pathway |
| WANG LMO4 TARGETS DN | 352 | 225 | All SZGR 2.0 genes in this pathway |
| AKL HTLV1 INFECTION DN | 68 | 41 | All SZGR 2.0 genes in this pathway |
| MULLIGHAN MLL SIGNATURE 2 UP | 418 | 263 | All SZGR 2.0 genes in this pathway |
| NAGASHIMA NRG1 SIGNALING DN | 58 | 35 | All SZGR 2.0 genes in this pathway |
| RODRIGUES THYROID CARCINOMA POORLY DIFFERENTIATED DN | 805 | 505 | All SZGR 2.0 genes in this pathway |
| KOKKINAKIS METHIONINE DEPRIVATION 48HR DN | 64 | 39 | All SZGR 2.0 genes in this pathway |
| KOKKINAKIS METHIONINE DEPRIVATION 96HR DN | 75 | 50 | All SZGR 2.0 genes in this pathway |
| MCBRYAN PUBERTAL BREAST 5 6WK DN | 137 | 97 | All SZGR 2.0 genes in this pathway |
| MCBRYAN PUBERTAL BREAST 6 7WK UP | 197 | 135 | All SZGR 2.0 genes in this pathway |
| PEREZ TP53 TARGETS | 1174 | 695 | All SZGR 2.0 genes in this pathway |
| APPIERTO RESPONSE TO FENRETINIDE UP | 38 | 26 | All SZGR 2.0 genes in this pathway |
| FARMER BREAST CANCER APOCRINE VS BASAL | 330 | 217 | All SZGR 2.0 genes in this pathway |
| DACOSTA UV RESPONSE VIA ERCC3 DN | 855 | 609 | All SZGR 2.0 genes in this pathway |
| DACOSTA UV RESPONSE VIA ERCC3 COMMON DN | 483 | 336 | All SZGR 2.0 genes in this pathway |
| BUYTAERT PHOTODYNAMIC THERAPY STRESS UP | 811 | 508 | All SZGR 2.0 genes in this pathway |
| LOPEZ MBD TARGETS | 957 | 597 | All SZGR 2.0 genes in this pathway |
| KOYAMA SEMA3B TARGETS DN | 411 | 249 | All SZGR 2.0 genes in this pathway |
| BENPORATH MYC MAX TARGETS | 775 | 494 | All SZGR 2.0 genes in this pathway |
| NIKOLSKY BREAST CANCER 7Q21 Q22 AMPLICON | 76 | 33 | All SZGR 2.0 genes in this pathway |
| NIKOLSKY MUTATED AND AMPLIFIED IN BREAST CANCER | 94 | 60 | All SZGR 2.0 genes in this pathway |
| DING LUNG CANCER EXPRESSION BY COPY NUMBER | 100 | 62 | All SZGR 2.0 genes in this pathway |
| NING CHRONIC OBSTRUCTIVE PULMONARY DISEASE DN | 121 | 79 | All SZGR 2.0 genes in this pathway |
| SHEPARD CRUSH AND BURN MUTANT DN | 185 | 111 | All SZGR 2.0 genes in this pathway |
| MATSUDA NATURAL KILLER DIFFERENTIATION | 475 | 313 | All SZGR 2.0 genes in this pathway |
| IVANOVA HEMATOPOIESIS EARLY PROGENITOR | 532 | 309 | All SZGR 2.0 genes in this pathway |
| BLALOCK ALZHEIMERS DISEASE INCIPIENT UP | 390 | 242 | All SZGR 2.0 genes in this pathway |
| IVANOVA HEMATOPOIESIS STEM CELL LONG TERM | 302 | 191 | All SZGR 2.0 genes in this pathway |
| KAYO AGING MUSCLE UP | 244 | 165 | All SZGR 2.0 genes in this pathway |
| BLALOCK ALZHEIMERS DISEASE UP | 1691 | 1088 | All SZGR 2.0 genes in this pathway |
| DAZARD RESPONSE TO UV NHEK DN | 318 | 220 | All SZGR 2.0 genes in this pathway |
| ZHANG BREAST CANCER PROGENITORS UP | 425 | 253 | All SZGR 2.0 genes in this pathway |
| ICHIBA GRAFT VERSUS HOST DISEASE D7 UP | 107 | 67 | All SZGR 2.0 genes in this pathway |
| TAVAZOIE METASTASIS | 108 | 68 | All SZGR 2.0 genes in this pathway |
| DANG BOUND BY MYC | 1103 | 714 | All SZGR 2.0 genes in this pathway |
| YAO TEMPORAL RESPONSE TO PROGESTERONE CLUSTER 2 | 86 | 50 | All SZGR 2.0 genes in this pathway |
| IKEDA MIR133 TARGETS UP | 43 | 27 | All SZGR 2.0 genes in this pathway |
| PLASARI TGFB1 SIGNALING VIA NFIC 1HR DN | 106 | 77 | All SZGR 2.0 genes in this pathway |
| FOSTER KDM1A TARGETS DN | 211 | 119 | All SZGR 2.0 genes in this pathway |
| ZWANG TRANSIENTLY UP BY 1ST EGF PULSE ONLY | 1839 | 928 | All SZGR 2.0 genes in this pathway |
| ZWANG EGF INTERVAL DN | 214 | 124 | All SZGR 2.0 genes in this pathway |