Gene Page: GPHN
Summary ?
| GeneID | 10243 |
| Symbol | GPHN |
| Synonyms | GEPH|GPH|GPHRYN|HKPX1|MOCODC |
| Description | gephyrin |
| Reference | MIM:603930|HGNC:HGNC:15465|Ensembl:ENSG00000171723|HPRD:04893|Vega:OTTHUMG00000029785 |
| Gene type | protein-coding |
| Map location | 14q23.3 |
| Pascal p-value | 0.046 |
| Sherlock p-value | 0.021 |
| Fetal beta | 0.343 |
| eGene | Cerebellar Hemisphere Myers' cis & trans |
| Support | G2Cdb.human_BAYES-COLLINS-HUMAN-PSD-CONSENSUS G2Cdb.human_BAYES-COLLINS-HUMAN-PSD-FULL G2Cdb.human_BAYES-COLLINS-MOUSE-PSD-CONSENSUS G2Cdb.humanPSD G2Cdb.humanPSP CompositeSet |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CV:PGCnp | Genome-wide Association Study | GWAS | |
| PMID:cooccur | High-throughput literature-search | Systematic search in PubMed for genes co-occurring with SCZ keywords. A total of 3027 genes were included. | |
| Expression | Meta-analysis of gene expression | P value: 2.648 | |
| Literature | High-throughput literature-search | Co-occurance with Schizophrenia keywords: schizophrenia,schizophrenias | Click to show details |
| GO_Annotation | Mapping neuro-related keywords to Gene Ontology annotations | Hits with neuro-related keywords: 2 |
Section I. Genetics and epigenetics annotation
 eQTL annotation
eQTL annotation
| SNP ID | Chromosome | Position | eGene | Gene Entrez ID | pvalue | qvalue | TSS distance | eQTL type |
|---|---|---|---|---|---|---|---|---|
| rs1020772 | chr9 | 76532511 | GPHN | 10243 | 0.11 | trans |
Section II. Transcriptome annotation
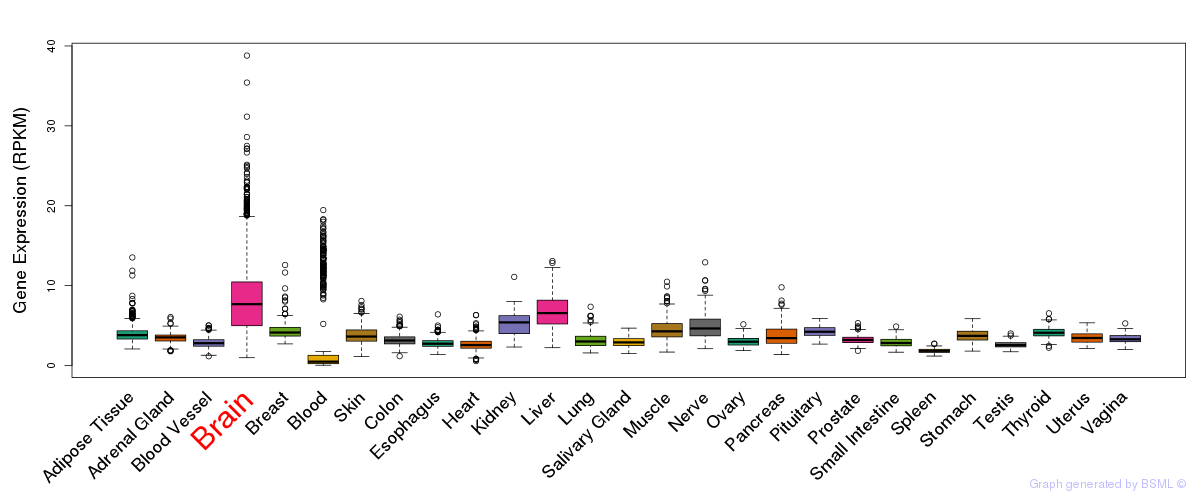
General gene expression (GTEx)
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
Top co-expressed genes in brain regions
| Top 10 positively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| SPTLC1 | 0.82 | 0.82 |
| C14orf100 | 0.81 | 0.83 |
| PAIP2 | 0.80 | 0.80 |
| C6orf211 | 0.80 | 0.81 |
| WDR51B | 0.79 | 0.76 |
| KIAA1279 | 0.79 | 0.80 |
| COG6 | 0.79 | 0.76 |
| USO1 | 0.78 | 0.73 |
| UBAP1 | 0.78 | 0.77 |
| ERGIC2 | 0.77 | 0.73 |
| Top 10 negatively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| IL32 | -0.50 | -0.37 |
| AF347015.18 | -0.49 | -0.36 |
| AC100783.1 | -0.44 | -0.37 |
| AC098691.2 | -0.44 | -0.32 |
| AF347015.21 | -0.44 | -0.23 |
| MT-CO2 | -0.42 | -0.28 |
| AC010300.1 | -0.41 | -0.39 |
| AF347015.2 | -0.41 | -0.27 |
| AF347015.26 | -0.40 | -0.27 |
| AF347015.8 | -0.39 | -0.26 |
Section III. Gene Ontology annotation
| Molecular function | GO term | Evidence | Neuro keywords | PubMed ID |
|---|---|---|---|---|
| GO:0000166 | nucleotide binding | IEA | - | |
| GO:0000287 | magnesium ion binding | IEA | - | |
| GO:0003824 | catalytic activity | IEA | - | |
| Biological process | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0007529 | establishment of synaptic specificity at neuromuscular junction | IEA | neuron, Synap (GO term level: 11) | - |
| GO:0006777 | Mo-molybdopterin cofactor biosynthetic process | IEA | - | |
| Cellular component | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0045211 | postsynaptic membrane | IEA | Synap, Neurotransmitter (GO term level: 5) | - |
| GO:0045202 | synapse | IEA | neuron, Synap, Neurotransmitter, Glial (GO term level: 2) | - |
| GO:0005856 | cytoskeleton | IEA | - | |
| GO:0005622 | intracellular | IEA | - | |
| GO:0005737 | cytoplasm | IEA | - | |
| GO:0005886 | plasma membrane | IEA | - | |
| GO:0030054 | cell junction | IEA | - |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| BIOCARTA GABA PATHWAY | 10 | 10 | All SZGR 2.0 genes in this pathway |
| REACTOME METABOLISM OF VITAMINS AND COFACTORS | 51 | 36 | All SZGR 2.0 genes in this pathway |
| PARENT MTOR SIGNALING UP | 567 | 375 | All SZGR 2.0 genes in this pathway |
| SENESE HDAC3 TARGETS UP | 501 | 327 | All SZGR 2.0 genes in this pathway |
| GRAESSMANN APOPTOSIS BY DOXORUBICIN DN | 1781 | 1082 | All SZGR 2.0 genes in this pathway |
| SPIELMAN LYMPHOBLAST EUROPEAN VS ASIAN DN | 584 | 395 | All SZGR 2.0 genes in this pathway |
| MORI SMALL PRE BII LYMPHOCYTE DN | 76 | 52 | All SZGR 2.0 genes in this pathway |
| IVANOVA HEMATOPOIESIS EARLY PROGENITOR | 532 | 309 | All SZGR 2.0 genes in this pathway |
| ULE SPLICING VIA NOVA2 | 43 | 38 | All SZGR 2.0 genes in this pathway |
| BURTON ADIPOGENESIS 5 | 126 | 78 | All SZGR 2.0 genes in this pathway |
| ZHENG BOUND BY FOXP3 | 491 | 310 | All SZGR 2.0 genes in this pathway |
| ZHENG FOXP3 TARGETS IN THYMUS UP | 196 | 137 | All SZGR 2.0 genes in this pathway |
| YAUCH HEDGEHOG SIGNALING PARACRINE UP | 149 | 85 | All SZGR 2.0 genes in this pathway |
| GRESHOCK CANCER COPY NUMBER UP | 323 | 240 | All SZGR 2.0 genes in this pathway |
| BOYLAN MULTIPLE MYELOMA D UP | 89 | 62 | All SZGR 2.0 genes in this pathway |
| YOSHIMURA MAPK8 TARGETS DN | 366 | 257 | All SZGR 2.0 genes in this pathway |
| HOFFMANN LARGE TO SMALL PRE BII LYMPHOCYTE UP | 163 | 102 | All SZGR 2.0 genes in this pathway |
| CHIANG LIVER CANCER SUBCLASS CTNNB1 UP | 176 | 110 | All SZGR 2.0 genes in this pathway |
| BOYLAN MULTIPLE MYELOMA PCA3 UP | 80 | 54 | All SZGR 2.0 genes in this pathway |
| WANG RESPONSE TO GSK3 INHIBITOR SB216763 UP | 397 | 206 | All SZGR 2.0 genes in this pathway |
| PILON KLF1 TARGETS DN | 1972 | 1213 | All SZGR 2.0 genes in this pathway |
| PURBEY TARGETS OF CTBP1 NOT SATB1 DN | 448 | 282 | All SZGR 2.0 genes in this pathway |
Section VI. microRNA annotation
| miRNA family | Target position | miRNA ID | miRNA seq | ||
|---|---|---|---|---|---|
| UTR start | UTR end | Match method | |||
| miR-138 | 742 | 748 | 1A | hsa-miR-138brain | AGCUGGUGUUGUGAAUC |
| miR-141/200a | 736 | 742 | 1A | hsa-miR-141 | UAACACUGUCUGGUAAAGAUGG |
| hsa-miR-200a | UAACACUGUCUGGUAACGAUGU | ||||
| miR-145 | 600 | 606 | m8 | hsa-miR-145 | GUCCAGUUUUCCCAGGAAUCCCUU |
| miR-153 | 61 | 67 | 1A | hsa-miR-153 | UUGCAUAGUCACAAAAGUGA |
| miR-190 | 36 | 43 | 1A,m8 | hsa-miR-190 | UGAUAUGUUUGAUAUAUUAGGU |
| miR-24* | 383 | 390 | 1A,m8 | hsa-miR-189 | GUGCCUACUGAGCUGAUAUCAGU |
| miR-299-3p | 34 | 40 | 1A | hsa-miR-299-3p | UAUGUGGGAUGGUAAACCGCUU |
| miR-330 | 240 | 246 | m8 | hsa-miR-330brain | GCAAAGCACACGGCCUGCAGAGA |
| miR-361 | 263 | 269 | m8 | hsa-miR-361brain | UUAUCAGAAUCUCCAGGGGUAC |
| miR-375 | 694 | 700 | 1A | hsa-miR-375 | UUUGUUCGUUCGGCUCGCGUGA |
| miR-448 | 60 | 67 | 1A,m8 | hsa-miR-448 | UUGCAUAUGUAGGAUGUCCCAU |
| miR-539 | 317 | 323 | 1A | hsa-miR-539 | GGAGAAAUUAUCCUUGGUGUGU |
| miR-543 | 780 | 786 | 1A | hsa-miR-543 | AAACAUUCGCGGUGCACUUCU |
| miR-544 | 302 | 308 | m8 | hsa-miR-544 | AUUCUGCAUUUUUAGCAAGU |
- SZ: miRNAs which differentially expressed in brain cortex of schizophrenia patients comparing with control samples using microarray. Click here to see the list of SZ related miRNAs.
- Brain: miRNAs which are expressed in brain based on miRNA microarray expression studies. Click here to see the list of brain related miRNAs.