Gene Page: EIF1B
Summary ?
| GeneID | 10289 |
| Symbol | EIF1B |
| Synonyms | GC20 |
| Description | eukaryotic translation initiation factor 1B |
| Reference | HGNC:HGNC:30792|Ensembl:ENSG00000114784|HPRD:17032|Vega:OTTHUMG00000131388 |
| Gene type | protein-coding |
| Map location | 3p22.1 |
| Pascal p-value | 0.223 |
| Sherlock p-value | 0.117 |
| Fetal beta | 0.526 |
| DMG | 1 (# studies) |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CV:PGCnp | Genome-wide Association Study | GWAS | |
| DMG:Jaffe_2016 | Genome-wide DNA methylation analysis | This dataset includes 2,104 probes/CpGs associated with SZ patients (n=108) compared to 136 controls at Bonferroni-adjusted P < 0.05. | 1 |
| Network | Shortest path distance of core genes in the Human protein-protein interaction network | Contribution to shortest path in PPI network: 0.059 |
Section I. Genetics and epigenetics annotation
 Differentially methylated gene
Differentially methylated gene
| Probe | Chromosome | Position | Nearest gene | P (dis) | Beta (dis) | FDR (dis) | Study |
|---|---|---|---|---|---|---|---|
| cg04019128 | 3 | 40350888 | EIF1B | 1.3E-9 | -0.012 | 1.35E-6 | DMG:Jaffe_2016 |
Section II. Transcriptome annotation
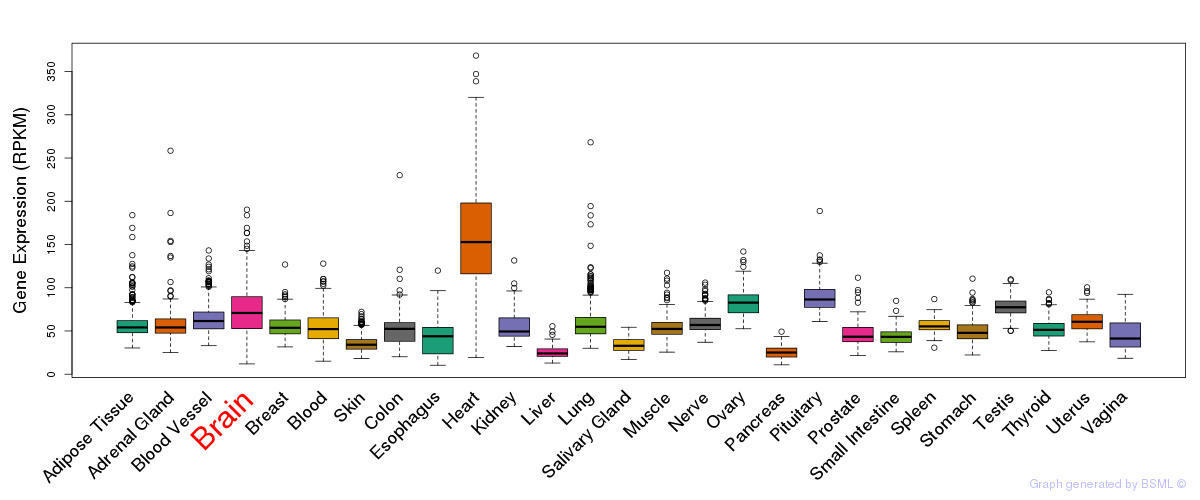
General gene expression (GTEx)
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
No co-expressed genes in brain regions
Section III. Gene Ontology annotation
| Molecular function | GO term | Evidence | Neuro keywords | PubMed ID |
|---|---|---|---|---|
| GO:0003743 | translation initiation factor activity | IEA | - | |
| GO:0005515 | protein binding | IPI | 17353931 | |
| Biological process | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0006446 | regulation of translational initiation | TAS | 7904817 |
Section IV. Protein-protein interaction annotation
| Interactors | Aliases B | Official full name B | Experimental | Source | PubMed ID |
|---|---|---|---|---|---|
| ACAT1 | ACAT | MAT | T2 | THIL | acetyl-Coenzyme A acetyltransferase 1 | Affinity Capture-MS | BioGRID | 17353931 |
| ACTR3 | ARP3 | ARP3 actin-related protein 3 homolog (yeast) | Affinity Capture-MS | BioGRID | 17353931 |
| EIF2S1 | EIF-2 | EIF-2A | EIF-2alpha | EIF2 | EIF2A | eukaryotic translation initiation factor 2, subunit 1 alpha, 35kDa | Affinity Capture-MS | BioGRID | 17353931 |
| EIF2S3 | EIF2 | EIF2G | EIF2gamma | eIF-2gA | eukaryotic translation initiation factor 2, subunit 3 gamma, 52kDa | Affinity Capture-MS | BioGRID | 17353931 |
| EIF3A | EIF3 | EIF3S10 | KIAA0139 | P167 | TIF32 | eIF3-p170 | eIF3-theta | p180 | p185 | eukaryotic translation initiation factor 3, subunit A | Affinity Capture-MS | BioGRID | 17353931 |
| EIF3D | EIF3S7 | MGC126526 | MGC17258 | eIF3-p66 | eIF3-zeta | eukaryotic translation initiation factor 3, subunit D | Affinity Capture-MS | BioGRID | 17353931 |
| EIF3E | EIF3-P48 | EIF3S6 | INT6 | eIF3-p46 | eukaryotic translation initiation factor 3, subunit E | Affinity Capture-MS | BioGRID | 17353931 |
| EIF3EIP | EIF3S11 | EIF3S6IP | HSPC021 | HSPC025 | MSTP005 | eukaryotic translation initiation factor 3, subunit E interacting protein | Affinity Capture-MS | BioGRID | 17353931 |
| EIF3F | EIF3S5 | eIF3-p47 | eukaryotic translation initiation factor 3, subunit F | Affinity Capture-MS | BioGRID | 17353931 |
| EIF3G | EIF3-P42 | EIF3S4 | eIF3-delta | eIF3-p44 | eukaryotic translation initiation factor 3, subunit G | Affinity Capture-MS | BioGRID | 17353931 |
| EIF3H | EIF3S3 | MGC102958 | eIF3-gamma | eIF3-p40 | eukaryotic translation initiation factor 3, subunit H | Affinity Capture-MS | BioGRID | 17353931 |
| EIF3I | EIF3S2 | PRO2242 | TRIP-1 | TRIP1 | eIF3-beta | eIF3-p36 | eukaryotic translation initiation factor 3, subunit I | Affinity Capture-MS | BioGRID | 17353931 |
| EIF3K | ARG134 | EIF3-p28 | EIF3S12 | HSPC029 | M9 | MSTP001 | PLAC-24 | PLAC24 | PRO1474 | PTD001 | eukaryotic translation initiation factor 3, subunit K | Affinity Capture-MS | BioGRID | 17353931 |
| EIF3M | B5 | FLJ29030 | GA17 | PCID1 | hfl-B5 | eukaryotic translation initiation factor 3, subunit M | Affinity Capture-MS | BioGRID | 17353931 |
| ERH | DROER | FLJ27340 | enhancer of rudimentary homolog (Drosophila) | Affinity Capture-MS | BioGRID | 17353931 |
| HPRT1 | HGPRT | HPRT | hypoxanthine phosphoribosyltransferase 1 | Affinity Capture-MS | BioGRID | 17353931 |
| MRCL3 | MLCB | MRLC3 | myosin regulatory light chain MRCL3 | Affinity Capture-MS | BioGRID | 17353931 |
| PCMT1 | - | protein-L-isoaspartate (D-aspartate) O-methyltransferase | Affinity Capture-MS | BioGRID | 17353931 |
| RAB7A | FLJ20819 | PRO2706 | RAB7 | RAB7A, member RAS oncogene family | Affinity Capture-MS | BioGRID | 17353931 |
| RPA3 | REPA3 | replication protein A3, 14kDa | Affinity Capture-MS | BioGRID | 17353931 |
| RPS11 | - | ribosomal protein S11 | Affinity Capture-MS | BioGRID | 17353931 |
| RPS29 | - | ribosomal protein S29 | Affinity Capture-MS | BioGRID | 17353931 |
| RPS3A | FTE1 | MFTL | MGC23240 | ribosomal protein S3A | Affinity Capture-MS | BioGRID | 17353931 |
| RPS9 | - | ribosomal protein S9 | Affinity Capture-MS | BioGRID | 17353931 |
| SHMT2 | GLYA | SHMT | serine hydroxymethyltransferase 2 (mitochondrial) | Affinity Capture-MS | BioGRID | 17353931 |
| TAGLN2 | HA1756 | KIAA0120 | transgelin 2 | Affinity Capture-MS | BioGRID | 17353931 |
| TCEB1 | SIII | transcription elongation factor B (SIII), polypeptide 1 (15kDa, elongin C) | Affinity Capture-MS | BioGRID | 17353931 |
| TSR1 | FLJ10534 | KIAA1401 | MGC131829 | TSR1, 20S rRNA accumulation, homolog (S. cerevisiae) | Affinity Capture-MS | BioGRID | 17353931 |
| YARS | CMTDIC | TYRRS | YRS | YTS | tyrosyl-tRNA synthetase | Affinity Capture-MS | BioGRID | 17353931 |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| ONKEN UVEAL MELANOMA DN | 526 | 357 | All SZGR 2.0 genes in this pathway |
| DIAZ CHRONIC MEYLOGENOUS LEUKEMIA UP | 1382 | 904 | All SZGR 2.0 genes in this pathway |
| TIEN INTESTINE PROBIOTICS 6HR DN | 167 | 100 | All SZGR 2.0 genes in this pathway |
| BIDUS METASTASIS DN | 161 | 93 | All SZGR 2.0 genes in this pathway |
| ENK UV RESPONSE KERATINOCYTE UP | 530 | 342 | All SZGR 2.0 genes in this pathway |
| LASTOWSKA NEUROBLASTOMA COPY NUMBER DN | 800 | 473 | All SZGR 2.0 genes in this pathway |
| GROSS HYPOXIA VIA HIF1A DN | 110 | 78 | All SZGR 2.0 genes in this pathway |
| NING CHRONIC OBSTRUCTIVE PULMONARY DISEASE DN | 121 | 79 | All SZGR 2.0 genes in this pathway |
| PENG GLUCOSE DEPRIVATION DN | 169 | 112 | All SZGR 2.0 genes in this pathway |
| BLALOCK ALZHEIMERS DISEASE DN | 1237 | 837 | All SZGR 2.0 genes in this pathway |
| HU GENOTOXIC DAMAGE 24HR | 35 | 22 | All SZGR 2.0 genes in this pathway |
| DAZARD RESPONSE TO UV NHEK UP | 244 | 151 | All SZGR 2.0 genes in this pathway |
| LEE CALORIE RESTRICTION MUSCLE DN | 51 | 28 | All SZGR 2.0 genes in this pathway |
| ACEVEDO LIVER CANCER UP | 973 | 570 | All SZGR 2.0 genes in this pathway |
| ACEVEDO LIVER TUMOR VS NORMAL ADJACENT TISSUE UP | 863 | 514 | All SZGR 2.0 genes in this pathway |
| BLUM RESPONSE TO SALIRASIB UP | 245 | 159 | All SZGR 2.0 genes in this pathway |
| ALONSO METASTASIS UP | 198 | 128 | All SZGR 2.0 genes in this pathway |
| YAO TEMPORAL RESPONSE TO PROGESTERONE CLUSTER 10 | 69 | 38 | All SZGR 2.0 genes in this pathway |
| HIRSCH CELLULAR TRANSFORMATION SIGNATURE UP | 242 | 159 | All SZGR 2.0 genes in this pathway |
| JOHNSTONE PARVB TARGETS 3 DN | 918 | 550 | All SZGR 2.0 genes in this pathway |
Section VI. microRNA annotation
| miRNA family | Target position | miRNA ID | miRNA seq | ||
|---|---|---|---|---|---|
| UTR start | UTR end | Match method | |||
| miR-299-5p | 304 | 310 | 1A | hsa-miR-299-5p | UGGUUUACCGUCCCACAUACAU |
| miR-494 | 223 | 229 | m8 | hsa-miR-494brain | UGAAACAUACACGGGAAACCUCUU |
- SZ: miRNAs which differentially expressed in brain cortex of schizophrenia patients comparing with control samples using microarray. Click here to see the list of SZ related miRNAs.
- Brain: miRNAs which are expressed in brain based on miRNA microarray expression studies. Click here to see the list of brain related miRNAs.