Gene Page: PPIE
Summary ?
| GeneID | 10450 |
| Symbol | PPIE |
| Synonyms | CYP-33|CYP33 |
| Description | peptidylprolyl isomerase E |
| Reference | MIM:602435|HGNC:HGNC:9258|Ensembl:ENSG00000084072|HPRD:03893|Vega:OTTHUMG00000009248 |
| Gene type | protein-coding |
| Map location | 1p32 |
| Pascal p-value | 0.228 |
| Sherlock p-value | 0.131 |
| Fetal beta | -0.168 |
| eGene | Caudate basal ganglia Cerebellar Hemisphere Cerebellum Cortex Frontal Cortex BA9 Hippocampus Hypothalamus Nucleus accumbens basal ganglia Putamen basal ganglia Meta |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CV:GWASdb | Genome-wide Association Studies | GWASdb records for schizophrenia | |
| CV:PGCnp | Genome-wide Association Study | GWAS |
Section I. Genetics and epigenetics annotation
 eQTL annotation
eQTL annotation
| SNP ID | Chromosome | Position | eGene | Gene Entrez ID | pvalue | qvalue | TSS distance | eQTL type |
|---|---|---|---|---|---|---|---|---|
| rs11206724 | 1 | 40200894 | PPIE | ENSG00000084072.12 | 4.3E-6 | 0 | 43040 | gtex_brain_putamen_basal |
| rs35895054 | 1 | 40202174 | PPIE | ENSG00000084072.12 | 4.613E-9 | 0 | 44320 | gtex_brain_putamen_basal |
| rs6658273 | 1 | 40202722 | PPIE | ENSG00000084072.12 | 5.261E-9 | 0 | 44868 | gtex_brain_putamen_basal |
| rs12074147 | 1 | 40203722 | PPIE | ENSG00000084072.12 | 2.022E-11 | 0 | 45868 | gtex_brain_putamen_basal |
| rs7513045 | 1 | 40204166 | PPIE | ENSG00000084072.12 | 4.925E-9 | 0 | 46312 | gtex_brain_putamen_basal |
| rs7548120 | 1 | 40204424 | PPIE | ENSG00000084072.12 | 2.022E-11 | 0 | 46570 | gtex_brain_putamen_basal |
| rs3738673 | 1 | 40204823 | PPIE | ENSG00000084072.12 | 2.022E-11 | 0 | 46969 | gtex_brain_putamen_basal |
| rs35698130 | 1 | 40205127 | PPIE | ENSG00000084072.12 | 2.022E-11 | 0 | 47273 | gtex_brain_putamen_basal |
| rs11809515 | 1 | 40205326 | PPIE | ENSG00000084072.12 | 1.319E-11 | 0 | 47472 | gtex_brain_putamen_basal |
| rs6664570 | 1 | 40206298 | PPIE | ENSG00000084072.12 | 2.022E-11 | 0 | 48444 | gtex_brain_putamen_basal |
| rs12086750 | 1 | 40210468 | PPIE | ENSG00000084072.12 | 2.022E-11 | 0 | 52614 | gtex_brain_putamen_basal |
| rs10888958 | 1 | 40211737 | PPIE | ENSG00000084072.12 | 2.022E-11 | 0 | 53883 | gtex_brain_putamen_basal |
| rs9787249 | 1 | 40212878 | PPIE | ENSG00000084072.12 | 2.022E-11 | 0 | 55024 | gtex_brain_putamen_basal |
| rs58589165 | 1 | 40213670 | PPIE | ENSG00000084072.12 | 2.072E-11 | 0 | 55816 | gtex_brain_putamen_basal |
| rs56064244 | 1 | 40213890 | PPIE | ENSG00000084072.12 | 2.072E-11 | 0 | 56036 | gtex_brain_putamen_basal |
| rs41267029 | 1 | 40214367 | PPIE | ENSG00000084072.12 | 2.072E-11 | 0 | 56513 | gtex_brain_putamen_basal |
| rs7520588 | 1 | 40214525 | PPIE | ENSG00000084072.12 | 2.072E-11 | 0 | 56671 | gtex_brain_putamen_basal |
| rs7518431 | 1 | 40214944 | PPIE | ENSG00000084072.12 | 2.072E-11 | 0 | 57090 | gtex_brain_putamen_basal |
| rs34701288 | 1 | 40215177 | PPIE | ENSG00000084072.12 | 3.253E-7 | 0 | 57323 | gtex_brain_putamen_basal |
| rs11206754 | 1 | 40216234 | PPIE | ENSG00000084072.12 | 2E-11 | 0 | 58380 | gtex_brain_putamen_basal |
| rs12239844 | 1 | 40217258 | PPIE | ENSG00000084072.12 | 2.022E-11 | 0 | 59404 | gtex_brain_putamen_basal |
| rs1046988 | 1 | 40219065 | PPIE | ENSG00000084072.12 | 2.022E-11 | 0 | 61211 | gtex_brain_putamen_basal |
| rs14528 | 1 | 40219280 | PPIE | ENSG00000084072.12 | 1.616E-9 | 0 | 61426 | gtex_brain_putamen_basal |
| rs6670841 | 1 | 40220284 | PPIE | ENSG00000084072.12 | 2.022E-11 | 0 | 62430 | gtex_brain_putamen_basal |
| rs7547787 | 1 | 40220806 | PPIE | ENSG00000084072.12 | 2.022E-11 | 0 | 62952 | gtex_brain_putamen_basal |
| rs2274949 | 1 | 40221067 | PPIE | ENSG00000084072.12 | 2.022E-11 | 0 | 63213 | gtex_brain_putamen_basal |
| rs1053846 | 1 | 40223140 | PPIE | ENSG00000084072.12 | 3.456E-7 | 0 | 65286 | gtex_brain_putamen_basal |
| rs12094291 | 1 | 40226046 | PPIE | ENSG00000084072.12 | 2.041E-11 | 0 | 68192 | gtex_brain_putamen_basal |
| rs28405581 | 1 | 40227057 | PPIE | ENSG00000084072.12 | 4.648E-7 | 0 | 69203 | gtex_brain_putamen_basal |
| rs2463260 | 1 | 40229368 | PPIE | ENSG00000084072.12 | 2.161E-7 | 0 | 71514 | gtex_brain_putamen_basal |
| rs2249439 | 1 | 40246491 | PPIE | ENSG00000084072.12 | 2.284E-8 | 0 | 88637 | gtex_brain_putamen_basal |
| rs1883648 | 1 | 40246535 | PPIE | ENSG00000084072.12 | 2.279E-8 | 0 | 88681 | gtex_brain_putamen_basal |
| rs1883649 | 1 | 40246549 | PPIE | ENSG00000084072.12 | 2.284E-8 | 0 | 88695 | gtex_brain_putamen_basal |
Section II. Transcriptome annotation
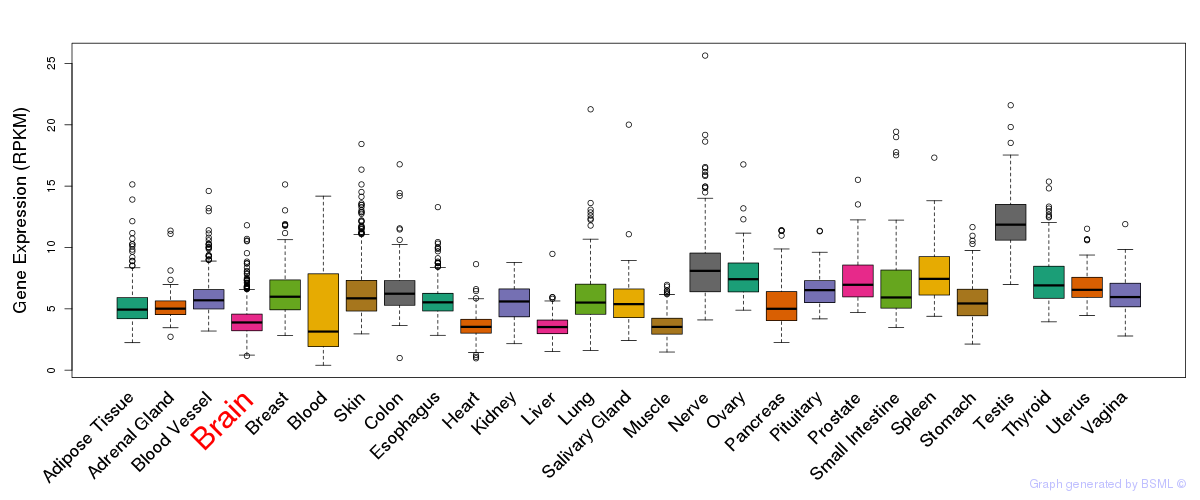
General gene expression (GTEx)
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
No co-expressed genes in brain regions
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| KEGG SPLICEOSOME | 128 | 72 | All SZGR 2.0 genes in this pathway |
| PARENT MTOR SIGNALING UP | 567 | 375 | All SZGR 2.0 genes in this pathway |
| ZHONG RESPONSE TO AZACITIDINE AND TSA DN | 70 | 38 | All SZGR 2.0 genes in this pathway |
| DIAZ CHRONIC MEYLOGENOUS LEUKEMIA UP | 1382 | 904 | All SZGR 2.0 genes in this pathway |
| LASTOWSKA NEUROBLASTOMA COPY NUMBER DN | 800 | 473 | All SZGR 2.0 genes in this pathway |
| DACOSTA UV RESPONSE VIA ERCC3 UP | 309 | 199 | All SZGR 2.0 genes in this pathway |
| SPIELMAN LYMPHOBLAST EUROPEAN VS ASIAN DN | 584 | 395 | All SZGR 2.0 genes in this pathway |
| LOPEZ MBD TARGETS | 957 | 597 | All SZGR 2.0 genes in this pathway |
| HU GENOTOXIN ACTION DIRECT VS INDIRECT 4HR | 37 | 22 | All SZGR 2.0 genes in this pathway |
| PAL PRMT5 TARGETS DN | 29 | 16 | All SZGR 2.0 genes in this pathway |
| MARSON BOUND BY FOXP3 STIMULATED | 1022 | 619 | All SZGR 2.0 genes in this pathway |
| BLUM RESPONSE TO SALIRASIB UP | 245 | 159 | All SZGR 2.0 genes in this pathway |
| DANG REGULATED BY MYC DN | 253 | 192 | All SZGR 2.0 genes in this pathway |
| DANG BOUND BY MYC | 1103 | 714 | All SZGR 2.0 genes in this pathway |