Gene Page: ADARB2
Summary ?
| GeneID | 105 |
| Symbol | ADARB2 |
| Synonyms | ADAR3|RED2 |
| Description | adenosine deaminase, RNA specific B2 (inactive) |
| Reference | MIM:602065|HGNC:HGNC:227|Ensembl:ENSG00000185736|HPRD:09070|Vega:OTTHUMG00000017543 |
| Gene type | protein-coding |
| Map location | 10p15.3 |
| Pascal p-value | 0.687 |
| Sherlock p-value | 0.218 |
| DMG | 2 (# studies) |
| eGene | Cerebellum Cortex Meta |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CV:PGCnp | Genome-wide Association Study | GWAS | |
| DMG:Jaffe_2016 | Genome-wide DNA methylation analysis | This dataset includes 2,104 probes/CpGs associated with SZ patients (n=108) compared to 136 controls at Bonferroni-adjusted P < 0.05. | 8 |
| DMG:Wockner_2014 | Genome-wide DNA methylation analysis | This dataset includes 4641 differentially methylated probes corresponding to 2929 unique genes between schizophrenia patients (n=24) and controls (n=24). | 8 |
Section I. Genetics and epigenetics annotation
 Differentially methylated gene
Differentially methylated gene
| Probe | Chromosome | Position | Nearest gene | P (dis) | Beta (dis) | FDR (dis) | Study |
|---|---|---|---|---|---|---|---|
| cg13504411 | 10 | 1596029 | ADARB2 | 4.19E-5 | 0.637 | 0.021 | DMG:Wockner_2014 |
| cg19163058 | 10 | 1595785 | ADARB2 | 4.54E-5 | 0.522 | 0.021 | DMG:Wockner_2014 |
| cg19533322 | 10 | 1595704 | ADARB2 | 7.36E-5 | 0.546 | 0.025 | DMG:Wockner_2014 |
| cg20731819 | 10 | 1509859 | ADARB2 | 1.043E-4 | 0.539 | 0.028 | DMG:Wockner_2014 |
| cg09265586 | 10 | 1595595 | ADARB2 | 3.557E-4 | 0.518 | 0.042 | DMG:Wockner_2014 |
| cg17898884 | 10 | 1725190 | ADARB2 | 4.249E-4 | -0.603 | 0.045 | DMG:Wockner_2014 |
| cg05289540 | 10 | 1595989 | ADARB2 | 5.013E-4 | 0.449 | 0.047 | DMG:Wockner_2014 |
| cg12557867 | 10 | 1779033 | ADARB2 | 9.89E-8 | -0.007 | 2.2E-5 | DMG:Jaffe_2016 |
Section II. Transcriptome annotation
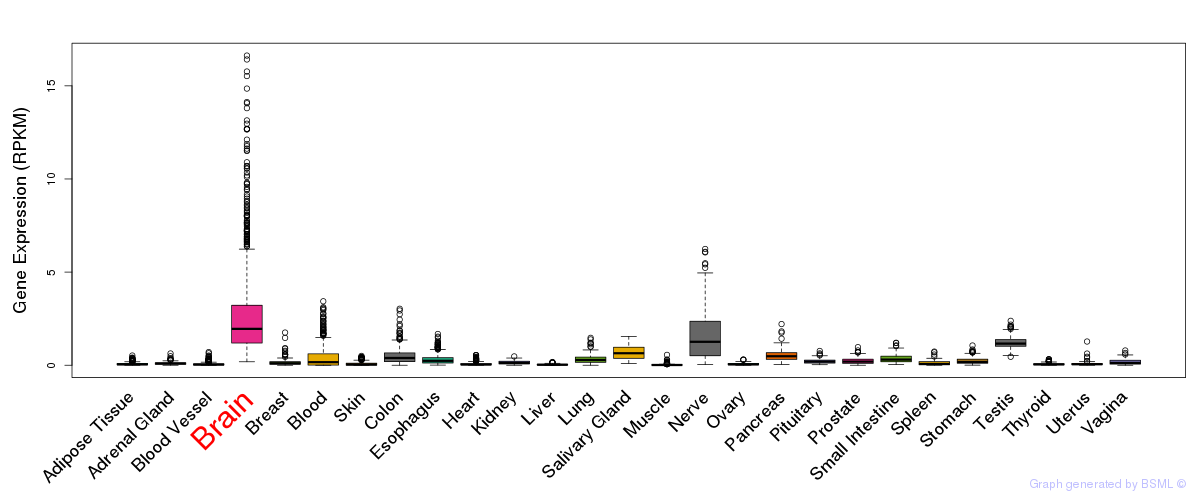
General gene expression (GTEx)
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
Top co-expressed genes in brain regions
| Top 10 positively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| SUSD5 | 0.83 | 0.78 |
| SIRPA | 0.83 | 0.80 |
| LRRK1 | 0.82 | 0.75 |
| PACSIN1 | 0.81 | 0.75 |
| IQSEC2 | 0.80 | 0.73 |
| MICAL2 | 0.80 | 0.76 |
| RASGRF2 | 0.80 | 0.77 |
| CAMK2A | 0.80 | 0.72 |
| PDE8B | 0.79 | 0.81 |
| PIP5K1B | 0.79 | 0.78 |
| Top 10 negatively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| BCL7C | -0.52 | -0.59 |
| EXOSC8 | -0.49 | -0.47 |
| HEBP2 | -0.49 | -0.59 |
| SNHG12 | -0.48 | -0.52 |
| C9orf46 | -0.47 | -0.50 |
| RBMX2 | -0.47 | -0.47 |
| C21orf57 | -0.47 | -0.50 |
| RPS13P2 | -0.47 | -0.50 |
| RPL23A | -0.47 | -0.50 |
| LEMD1 | -0.45 | -0.35 |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| DODD NASOPHARYNGEAL CARCINOMA UP | 1821 | 933 | All SZGR 2.0 genes in this pathway |
| TANAKA METHYLATED IN ESOPHAGEAL CARCINOMA | 103 | 58 | All SZGR 2.0 genes in this pathway |
| SCHAEFFER PROSTATE DEVELOPMENT 48HR UP | 487 | 286 | All SZGR 2.0 genes in this pathway |
| BENPORATH SUZ12 TARGETS | 1038 | 678 | All SZGR 2.0 genes in this pathway |
| BENPORATH EED TARGETS | 1062 | 725 | All SZGR 2.0 genes in this pathway |
| BENPORATH ES WITH H3K27ME3 | 1118 | 744 | All SZGR 2.0 genes in this pathway |
| BENPORATH PRC2 TARGETS | 652 | 441 | All SZGR 2.0 genes in this pathway |
| SHEPARD CRUSH AND BURN MUTANT DN | 185 | 111 | All SZGR 2.0 genes in this pathway |
| BLALOCK ALZHEIMERS DISEASE UP | 1691 | 1088 | All SZGR 2.0 genes in this pathway |
| ACEVEDO METHYLATED IN LIVER CANCER DN | 940 | 425 | All SZGR 2.0 genes in this pathway |
| MIKKELSEN IPS ICP WITH H3K4ME3 AND H327ME3 | 126 | 83 | All SZGR 2.0 genes in this pathway |
| MIKKELSEN ES ICP WITH H3K4ME3 AND H3K27ME3 | 137 | 85 | All SZGR 2.0 genes in this pathway |
| DEMAGALHAES AGING UP | 55 | 39 | All SZGR 2.0 genes in this pathway |
| DUAN PRDM5 TARGETS | 79 | 52 | All SZGR 2.0 genes in this pathway |