Gene Page: ZNF211
Summary ?
| GeneID | 10520 |
| Symbol | ZNF211 |
| Synonyms | C2H2-25|CH2H2-25|ZNF-25|ZNFC25 |
| Description | zinc finger protein 211 |
| Reference | MIM:601856|HGNC:HGNC:13003|Ensembl:ENSG00000121417|HPRD:03512|Vega:OTTHUMG00000168012 |
| Gene type | protein-coding |
| Map location | 19q13.4 |
| Pascal p-value | 0.503 |
| Sherlock p-value | 0.868 |
| DMG | 1 (# studies) |
| eGene | Myers' cis & trans |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CV:PGCnp | Genome-wide Association Study | GWAS | |
| DMG:Wockner_2014 | Genome-wide DNA methylation analysis | This dataset includes 4641 differentially methylated probes corresponding to 2929 unique genes between schizophrenia patients (n=24) and controls (n=24). | 1 |
Section I. Genetics and epigenetics annotation
 Differentially methylated gene
Differentially methylated gene
| Probe | Chromosome | Position | Nearest gene | P (dis) | Beta (dis) | FDR (dis) | Study |
|---|---|---|---|---|---|---|---|
| cg25477163 | 19 | 58147078 | ZNF211 | 9.41E-6 | 0.569 | 0.013 | DMG:Wockner_2014 |
 eQTL annotation
eQTL annotation
| SNP ID | Chromosome | Position | eGene | Gene Entrez ID | pvalue | qvalue | TSS distance | eQTL type |
|---|---|---|---|---|---|---|---|---|
| rs2079033 | chr19 | 58129457 | ZNF211 | 10520 | 0.08 | cis | ||
| rs9749085 | chr19 | 58142485 | ZNF211 | 10520 | 0.13 | cis | ||
| rs3746209 | chr19 | 58154304 | ZNF211 | 10520 | 0.07 | cis | ||
| rs10408464 | chr19 | 58154824 | ZNF211 | 10520 | 0.05 | cis | ||
| rs10409262 | chr19 | 58154954 | ZNF211 | 10520 | 0.08 | cis | ||
| rs459934 | chr5 | 132847093 | ZNF211 | 10520 | 0.11 | trans |
Section II. Transcriptome annotation
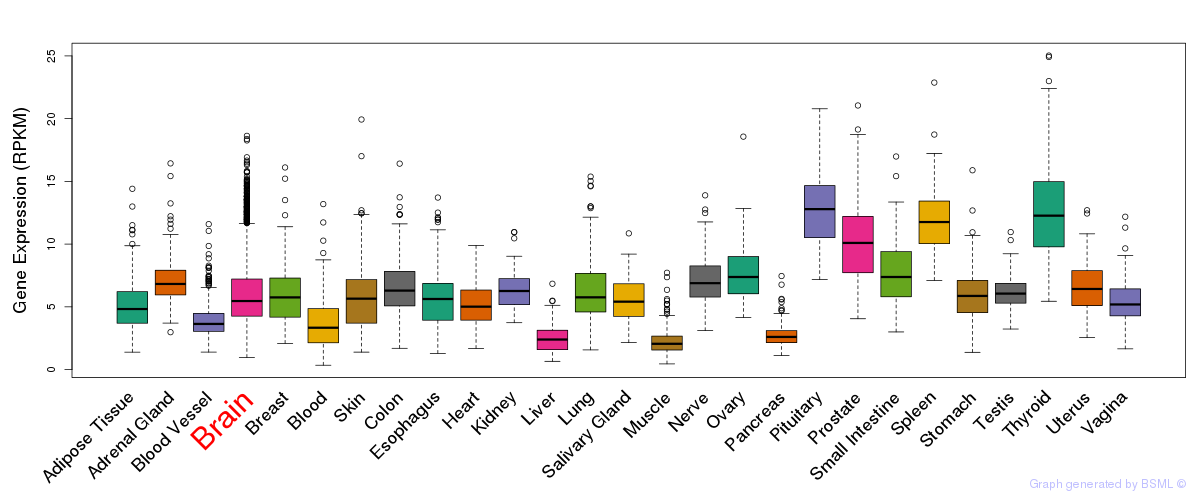
General gene expression (GTEx)
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
Top co-expressed genes in brain regions
| Top 10 positively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| EIF3I | 0.89 | 0.87 |
| CCT7 | 0.86 | 0.84 |
| SNRPC | 0.86 | 0.81 |
| PRMT5 | 0.85 | 0.81 |
| PSMA7 | 0.85 | 0.83 |
| PHF5A | 0.84 | 0.80 |
| WDR25 | 0.84 | 0.83 |
| COPZ1 | 0.84 | 0.82 |
| RAN | 0.84 | 0.81 |
| H2AFZ | 0.83 | 0.82 |
| Top 10 negatively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| AF347015.8 | -0.61 | -0.63 |
| AF347015.26 | -0.61 | -0.66 |
| MT-CO2 | -0.61 | -0.61 |
| AF347015.27 | -0.60 | -0.64 |
| AF347015.2 | -0.60 | -0.64 |
| AF347015.33 | -0.60 | -0.63 |
| AF347015.15 | -0.60 | -0.63 |
| MT-CYB | -0.60 | -0.62 |
| AF347015.31 | -0.58 | -0.60 |
| AF347015.18 | -0.56 | -0.67 |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| REACTOME GENERIC TRANSCRIPTION PATHWAY | 352 | 181 | All SZGR 2.0 genes in this pathway |
| ENK UV RESPONSE KERATINOCYTE UP | 530 | 342 | All SZGR 2.0 genes in this pathway |
| AMUNDSON GENOTOXIC SIGNATURE | 105 | 68 | All SZGR 2.0 genes in this pathway |
| JAZAERI BREAST CANCER BRCA1 VS BRCA2 DN | 43 | 31 | All SZGR 2.0 genes in this pathway |
| BLALOCK ALZHEIMERS DISEASE UP | 1691 | 1088 | All SZGR 2.0 genes in this pathway |
| KAYO CALORIE RESTRICTION MUSCLE UP | 95 | 64 | All SZGR 2.0 genes in this pathway |
| KRIGE RESPONSE TO TOSEDOSTAT 6HR UP | 953 | 554 | All SZGR 2.0 genes in this pathway |
| KRIGE RESPONSE TO TOSEDOSTAT 24HR UP | 783 | 442 | All SZGR 2.0 genes in this pathway |
| AGUIRRE PANCREATIC CANCER COPY NUMBER UP | 298 | 174 | All SZGR 2.0 genes in this pathway |
| JISON SICKLE CELL DISEASE DN | 181 | 97 | All SZGR 2.0 genes in this pathway |
| VERHAAK GLIOBLASTOMA CLASSICAL | 162 | 122 | All SZGR 2.0 genes in this pathway |
| ZWANG TRANSIENTLY UP BY 1ST EGF PULSE ONLY | 1839 | 928 | All SZGR 2.0 genes in this pathway |