Gene Page: DDX17
Summary ?
| GeneID | 10521 |
| Symbol | DDX17 |
| Synonyms | P72|RH70 |
| Description | DEAD-box helicase 17 |
| Reference | MIM:608469|HGNC:HGNC:2740|Ensembl:ENSG00000100201|HPRD:10532|Vega:OTTHUMG00000151136 |
| Gene type | protein-coding |
| Map location | 22q13.1 |
| Pascal p-value | 0.027 |
| Sherlock p-value | 4.424E-6 |
| Fetal beta | 1.747 |
| DMG | 1 (# studies) |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CV:PGCnp | Genome-wide Association Study | GWAS | |
| DMG:Nishioka_2013 | Genome-wide DNA methylation analysis | The authors investigated the methylation profiles of DNA in peripheral blood cells from 18 patients with first-episode schizophrenia (FESZ) and from 15 normal controls. | 1 |
Section I. Genetics and epigenetics annotation
 Differentially methylated gene
Differentially methylated gene
| Probe | Chromosome | Position | Nearest gene | P (dis) | Beta (dis) | FDR (dis) | Study |
|---|---|---|---|---|---|---|---|
| cg03640568 | 22 | 38902623 | DDX17 | -0.016 | 0.98 | DMG:Nishioka_2013 |
Section II. Transcriptome annotation
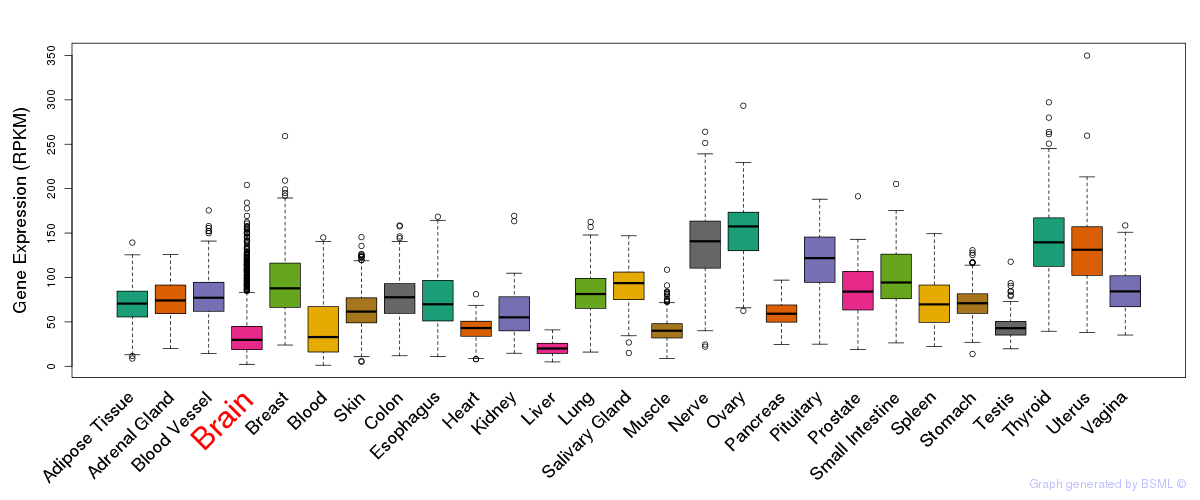
General gene expression (GTEx)
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
Top co-expressed genes in brain regions
| Top 10 positively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| PXMP3 | 0.92 | 0.82 |
| SCRG1 | 0.86 | 0.77 |
| GSTO1 | 0.85 | 0.83 |
| LPAR6 | 0.84 | 0.74 |
| HSD17B8 | 0.83 | 0.67 |
| MT3 | 0.83 | 0.76 |
| LY96 | 0.83 | 0.72 |
| HOPX | 0.82 | 0.76 |
| C2orf7 | 0.81 | 0.69 |
| MRPS15 | 0.81 | 0.68 |
| Top 10 negatively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| TNKS1BP1 | -0.63 | -0.73 |
| MYO18A | -0.61 | -0.61 |
| SUPT6H | -0.60 | -0.63 |
| PHRF1 | -0.59 | -0.71 |
| CDC2L2 | -0.59 | -0.63 |
| SAFB2 | -0.59 | -0.65 |
| ABCF1 | -0.58 | -0.62 |
| SMG6 | -0.58 | -0.66 |
| SART1 | -0.58 | -0.66 |
| GIGYF2 | -0.58 | -0.62 |
Section IV. Protein-protein interaction annotation
| Interactors | Aliases B | Official full name B | Experimental | Source | PubMed ID |
|---|---|---|---|---|---|
| DDX5 | DKFZp686J01190 | G17P1 | HLR1 | HUMP68 | p68 | DEAD (Asp-Glu-Ala-Asp) box polypeptide 5 | p72 interacts with p68. | BIND | 12595555 |
| ESR1 | DKFZp686N23123 | ER | ESR | ESRA | Era | NR3A1 | estrogen receptor 1 | ESR1 (ER-alpha) interacts with an unspecified isoform of DDX17 (p72). This interaction was modeled on a demonstrated interaction between ESR1 from an unspecified species and DDX17 from an unspecified species. | BIND | 12738788 |
| HDAC1 | DKFZp686H12203 | GON-10 | HD1 | RPD3 | RPD3L1 | histone deacetylase 1 | - | HPRD,BioGRID | 15298701 |
| LNX1 | LNX | MPDZ | PDZRN2 | ligand of numb-protein X 1 | Two-hybrid | BioGRID | 16189514 |
| NCOA1 | F-SRC-1 | KAT13A | MGC129719 | MGC129720 | NCoA-1 | RIP160 | SRC-1 | SRC1 | bHLHe42 | nuclear receptor coactivator 1 | - | HPRD,BioGRID | 11250900 |
| PCNA | MGC8367 | proliferating cell nuclear antigen | DDX17 is a PCNA-binding protein | BIND | 12171929 |
| SF1 | D11S636 | ZFM1 | ZNF162 | splicing factor 1 | Two-hybrid | BioGRID | 16189514 |
| SFRS12 | DKFZp564B176 | MGC133045 | SRrp508 | SRrp86 | splicing factor, arginine/serine-rich 12 | - | HPRD | 14559993 |
| SNRNP70 | RNPU1Z | RPU1 | SNRP70 | U170K | U1AP | U1RNP | small nuclear ribonucleoprotein 70kDa (U1) | - | HPRD,BioGRID | 12193588 |
| SORBS3 | SCAM-1 | SCAM1 | SH3D4 | VINEXIN | sorbin and SH3 domain containing 3 | Two-hybrid | BioGRID | 16189514 |
| WBP11 | DKFZp779M1063 | NPWBP | SIPP1 | WW domain binding protein 11 | Two-hybrid | BioGRID | 16189514 |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| PID ERA GENOMIC PATHWAY | 65 | 37 | All SZGR 2.0 genes in this pathway |
| SENGUPTA NASOPHARYNGEAL CARCINOMA WITH LMP1 UP | 408 | 247 | All SZGR 2.0 genes in this pathway |
| CHANDRAN METASTASIS TOP50 UP | 38 | 27 | All SZGR 2.0 genes in this pathway |
| OSWALD HEMATOPOIETIC STEM CELL IN COLLAGEN GEL DN | 275 | 168 | All SZGR 2.0 genes in this pathway |
| BOGNI TREATMENT RELATED MYELOID LEUKEMIA DN | 33 | 19 | All SZGR 2.0 genes in this pathway |
| RHEIN ALL GLUCOCORTICOID THERAPY UP | 78 | 41 | All SZGR 2.0 genes in this pathway |
| GRAHAM NORMAL QUIESCENT VS NORMAL DIVIDING UP | 66 | 47 | All SZGR 2.0 genes in this pathway |
| ENK UV RESPONSE KERATINOCYTE DN | 485 | 334 | All SZGR 2.0 genes in this pathway |
| GRAESSMANN APOPTOSIS BY DOXORUBICIN DN | 1781 | 1082 | All SZGR 2.0 genes in this pathway |
| MATTIOLI MGUS VS MULTIPLE MYELOMA | 16 | 11 | All SZGR 2.0 genes in this pathway |
| DACOSTA UV RESPONSE VIA ERCC3 DN | 855 | 609 | All SZGR 2.0 genes in this pathway |
| MARTORIATI MDM4 TARGETS FETAL LIVER DN | 514 | 319 | All SZGR 2.0 genes in this pathway |
| LIU NASOPHARYNGEAL CARCINOMA | 70 | 38 | All SZGR 2.0 genes in this pathway |
| WEI MIR34A TARGETS | 148 | 97 | All SZGR 2.0 genes in this pathway |
| SCIAN INVERSED TARGETS OF TP53 AND TP73 DN | 31 | 23 | All SZGR 2.0 genes in this pathway |
| RICKMAN TUMOR DIFFERENTIATED WELL VS POORLY UP | 236 | 139 | All SZGR 2.0 genes in this pathway |
| BENPORATH NANOG TARGETS | 988 | 594 | All SZGR 2.0 genes in this pathway |
| BENPORATH SOX2 TARGETS | 734 | 436 | All SZGR 2.0 genes in this pathway |
| BENPORATH MYC MAX TARGETS | 775 | 494 | All SZGR 2.0 genes in this pathway |
| SHEPARD BMYB MORPHOLINO DN | 200 | 112 | All SZGR 2.0 genes in this pathway |
| DER IFN ALPHA RESPONSE UP | 74 | 48 | All SZGR 2.0 genes in this pathway |
| SHEPARD BMYB TARGETS | 74 | 41 | All SZGR 2.0 genes in this pathway |
| SHEPARD CRUSH AND BURN MUTANT DN | 185 | 111 | All SZGR 2.0 genes in this pathway |
| DER IFN BETA RESPONSE UP | 102 | 67 | All SZGR 2.0 genes in this pathway |
| IIZUKA LIVER CANCER EARLY RECURRENCE | 11 | 10 | All SZGR 2.0 genes in this pathway |
| HADDAD T LYMPHOCYTE AND NK PROGENITOR UP | 78 | 56 | All SZGR 2.0 genes in this pathway |
| GENTILE UV RESPONSE CLUSTER D4 | 55 | 37 | All SZGR 2.0 genes in this pathway |
| KAYO CALORIE RESTRICTION MUSCLE DN | 87 | 59 | All SZGR 2.0 genes in this pathway |
| BROWNE HCMV INFECTION 24HR DN | 148 | 102 | All SZGR 2.0 genes in this pathway |
| BROWNE HCMV INFECTION 14HR DN | 298 | 200 | All SZGR 2.0 genes in this pathway |
| GENTILE UV HIGH DOSE DN | 312 | 203 | All SZGR 2.0 genes in this pathway |
| BAELDE DIABETIC NEPHROPATHY DN | 434 | 302 | All SZGR 2.0 genes in this pathway |
| BILD HRAS ONCOGENIC SIGNATURE | 261 | 166 | All SZGR 2.0 genes in this pathway |
| HELLER HDAC TARGETS DN | 292 | 189 | All SZGR 2.0 genes in this pathway |
| RIZKI TUMOR INVASIVENESS 3D DN | 270 | 181 | All SZGR 2.0 genes in this pathway |
| SMID BREAST CANCER BASAL UP | 648 | 398 | All SZGR 2.0 genes in this pathway |
| CHAUHAN RESPONSE TO METHOXYESTRADIOL DN | 102 | 65 | All SZGR 2.0 genes in this pathway |
| ZHANG TLX TARGETS 36HR UP | 221 | 150 | All SZGR 2.0 genes in this pathway |
| MELLMAN TUT1 TARGETS DN | 47 | 29 | All SZGR 2.0 genes in this pathway |
| KYNG WERNER SYNDROM AND NORMAL AGING DN | 225 | 124 | All SZGR 2.0 genes in this pathway |
| DANG BOUND BY MYC | 1103 | 714 | All SZGR 2.0 genes in this pathway |
| CHANDRAN METASTASIS UP | 221 | 135 | All SZGR 2.0 genes in this pathway |
| YAO TEMPORAL RESPONSE TO PROGESTERONE CLUSTER 1 | 68 | 50 | All SZGR 2.0 genes in this pathway |
| BROWNE HCMV INFECTION 30MIN DN | 150 | 99 | All SZGR 2.0 genes in this pathway |
| BROWNE HCMV INFECTION 4HR DN | 254 | 158 | All SZGR 2.0 genes in this pathway |
| PURBEY TARGETS OF CTBP1 AND SATB1 UP | 87 | 52 | All SZGR 2.0 genes in this pathway |
| ALFANO MYC TARGETS | 239 | 156 | All SZGR 2.0 genes in this pathway |
| RAO BOUND BY SALL4 ISOFORM B | 517 | 302 | All SZGR 2.0 genes in this pathway |
| ABRAMSON INTERACT WITH AIRE | 45 | 33 | All SZGR 2.0 genes in this pathway |
| ZWANG TRANSIENTLY UP BY 1ST EGF PULSE ONLY | 1839 | 928 | All SZGR 2.0 genes in this pathway |