Gene Page: CEL
Summary ?
| GeneID | 1056 |
| Symbol | CEL |
| Synonyms | BAL|BSDL|BSSL|CELL|CEase|FAP|FAPP|LIPA|MODY8 |
| Description | carboxyl ester lipase |
| Reference | MIM:114840|HGNC:HGNC:1848|Ensembl:ENSG00000170835|HPRD:07509|Vega:OTTHUMG00000020855 |
| Gene type | protein-coding |
| Map location | 9q34.3 |
| Pascal p-value | 0.139 |
| Fetal beta | 0.813 |
| eGene | Meta |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CV:PGCnp | Genome-wide Association Study | GWAS | |
| PMID:cooccur | High-throughput literature-search | Systematic search in PubMed for genes co-occurring with SCZ keywords. A total of 3027 genes were included. | |
| Literature | High-throughput literature-search | Co-occurance with Schizophrenia keywords: schizophrenia,schizophrenic,schizophrenias | Click to show details |
Section I. Genetics and epigenetics annotation
Section II. Transcriptome annotation
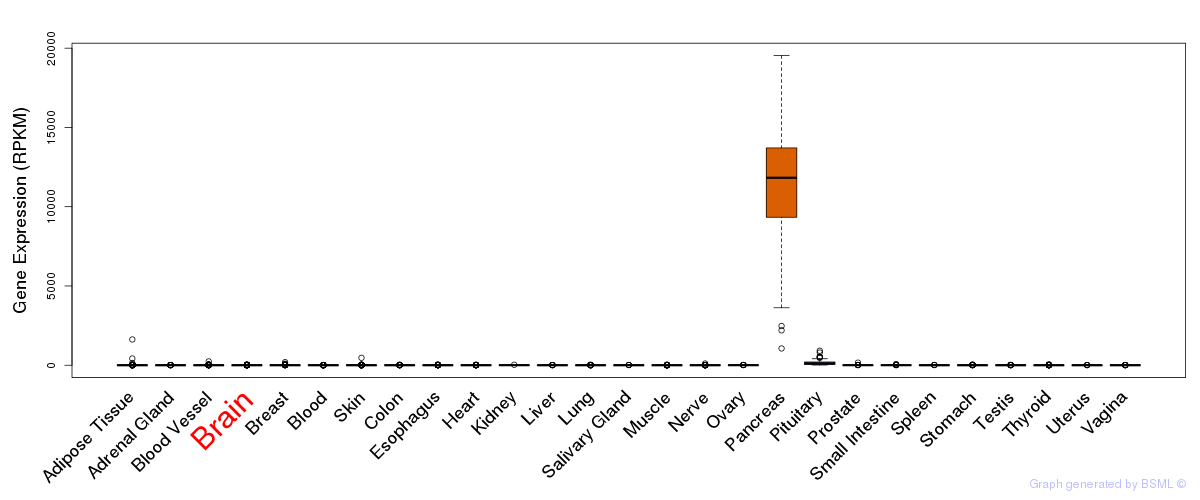
General gene expression (GTEx)
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
Top co-expressed genes in brain regions
| Top 10 positively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| MKI67 | 0.98 | 0.88 |
| KIF14 | 0.98 | 0.82 |
| ASPM | 0.97 | 0.82 |
| CASC5 | 0.96 | 0.85 |
| KIF15 | 0.96 | 0.85 |
| CENPE | 0.96 | 0.87 |
| MCM10 | 0.96 | 0.81 |
| TOP2A | 0.96 | 0.86 |
| TPX2 | 0.95 | 0.94 |
| POLQ | 0.95 | 0.80 |
| Top 10 negatively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| PTH1R | -0.41 | -0.76 |
| HLA-F | -0.40 | -0.81 |
| FBXO2 | -0.40 | -0.65 |
| C5orf53 | -0.40 | -0.75 |
| SLC9A3R2 | -0.40 | -0.48 |
| ALDOC | -0.39 | -0.73 |
| LHPP | -0.39 | -0.63 |
| AIFM3 | -0.39 | -0.77 |
| TNFSF12 | -0.39 | -0.68 |
| S100B | -0.38 | -0.84 |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| KEGG STEROID BIOSYNTHESIS | 17 | 12 | All SZGR 2.0 genes in this pathway |
| KEGG GLYCEROLIPID METABOLISM | 49 | 26 | All SZGR 2.0 genes in this pathway |
| WATANABE COLON CANCER MSI VS MSS DN | 81 | 42 | All SZGR 2.0 genes in this pathway |
| SENGUPTA NASOPHARYNGEAL CARCINOMA WITH LMP1 UP | 408 | 247 | All SZGR 2.0 genes in this pathway |
| GAUSSMANN MLL AF4 FUSION TARGETS G UP | 238 | 135 | All SZGR 2.0 genes in this pathway |
| PEREZ TP53 TARGETS | 1174 | 695 | All SZGR 2.0 genes in this pathway |
| RICKMAN HEAD AND NECK CANCER E | 89 | 44 | All SZGR 2.0 genes in this pathway |
| IGLESIAS E2F TARGETS DN | 16 | 11 | All SZGR 2.0 genes in this pathway |
| BROWNE HCMV INFECTION 48HR DN | 504 | 323 | All SZGR 2.0 genes in this pathway |
| BROWNE HCMV INFECTION 10HR UP | 101 | 69 | All SZGR 2.0 genes in this pathway |
| HELLER SILENCED BY METHYLATION UP | 282 | 183 | All SZGR 2.0 genes in this pathway |
| HELLER HDAC TARGETS SILENCED BY METHYLATION UP | 461 | 298 | All SZGR 2.0 genes in this pathway |
| HANN RESISTANCE TO BCL2 INHIBITOR DN | 48 | 31 | All SZGR 2.0 genes in this pathway |
| CROMER TUMORIGENESIS DN | 51 | 29 | All SZGR 2.0 genes in this pathway |
| BROWNE HCMV INFECTION 30MIN DN | 150 | 99 | All SZGR 2.0 genes in this pathway |
| BRUINS UVC RESPONSE VIA TP53 GROUP A | 898 | 516 | All SZGR 2.0 genes in this pathway |
| FEVR CTNNB1 TARGETS UP | 682 | 433 | All SZGR 2.0 genes in this pathway |
| PEDRIOLI MIR31 TARGETS DN | 418 | 245 | All SZGR 2.0 genes in this pathway |