Gene Page: MAP3K2
Summary ?
| GeneID | 10746 |
| Symbol | MAP3K2 |
| Synonyms | MEKK2|MEKK2B |
| Description | mitogen-activated protein kinase kinase kinase 2 |
| Reference | MIM:609487|HGNC:HGNC:6854|Ensembl:ENSG00000169967|HPRD:10068|Vega:OTTHUMG00000153397 |
| Gene type | protein-coding |
| Map location | 2q14.3 |
| Pascal p-value | 0.985 |
| Sherlock p-value | 0.023 |
| Fetal beta | 0.874 |
| eGene | Myers' cis & trans Meta |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| GSMA_I | Genome scan meta-analysis | Psr: 0.023 | |
| GSMA_IIA | Genome scan meta-analysis (All samples) | Psr: 0.00755 |
Section I. Genetics and epigenetics annotation
 eQTL annotation
eQTL annotation
| SNP ID | Chromosome | Position | eGene | Gene Entrez ID | pvalue | qvalue | TSS distance | eQTL type |
|---|---|---|---|---|---|---|---|---|
| rs2404535 | chr2 | 127947708 | MAP3K2 | 10746 | 0.14 | cis | ||
| rs7585198 | chr2 | 127962634 | MAP3K2 | 10746 | 0 | cis | ||
| rs12463909 | chr2 | 127971737 | MAP3K2 | 10746 | 0.01 | cis | ||
| rs4150477 | chr2 | 128032545 | MAP3K2 | 10746 | 0.04 | cis | ||
| rs6710535 | chr2 | 128190959 | MAP3K2 | 10746 | 0.03 | cis | ||
| rs7585198 | chr2 | 127962634 | MAP3K2 | 10746 | 0.13 | trans |
Section II. Transcriptome annotation
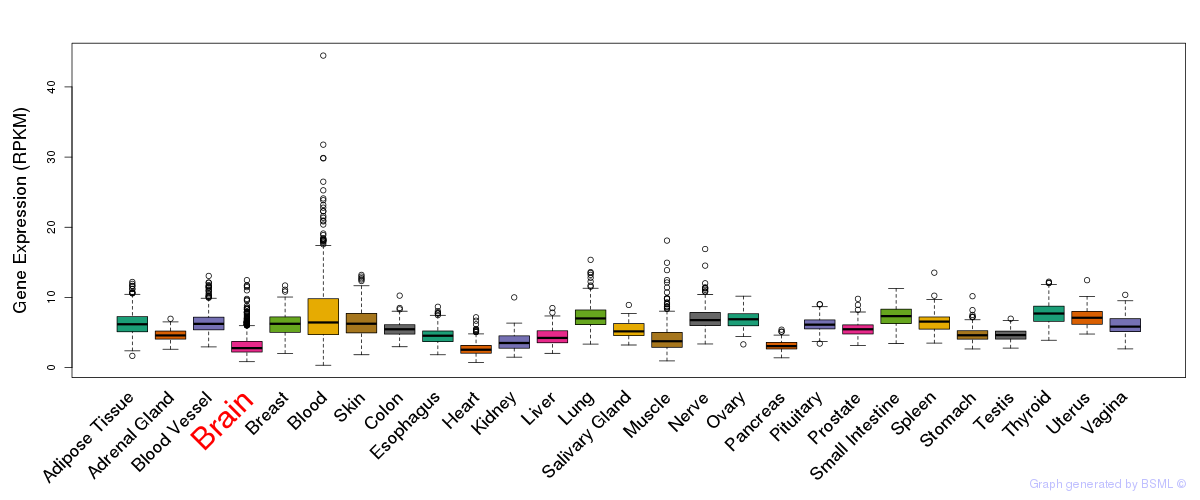
General gene expression (GTEx)
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
Top co-expressed genes in brain regions
| Top 10 positively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| PYGO2 | 0.85 | 0.87 |
| MAGED1 | 0.85 | 0.87 |
| KIAA1539 | 0.82 | 0.83 |
| AC069234.1 | 0.82 | 0.87 |
| ZMYM3 | 0.82 | 0.86 |
| VAC14 | 0.81 | 0.85 |
| C7orf43 | 0.81 | 0.85 |
| EFNB3 | 0.81 | 0.85 |
| LRRN2 | 0.81 | 0.85 |
| RGMB | 0.81 | 0.84 |
| Top 10 negatively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| MT-CO2 | -0.68 | -0.73 |
| AF347015.8 | -0.66 | -0.73 |
| AF347015.31 | -0.66 | -0.70 |
| AF347015.21 | -0.66 | -0.76 |
| AF347015.2 | -0.65 | -0.70 |
| MT-CYB | -0.64 | -0.68 |
| AF347015.33 | -0.63 | -0.68 |
| AF347015.27 | -0.63 | -0.69 |
| AF347015.15 | -0.62 | -0.67 |
| AF347015.26 | -0.62 | -0.68 |
Section III. Gene Ontology annotation
| Molecular function | GO term | Evidence | Neuro keywords | PubMed ID |
|---|---|---|---|---|
| GO:0000166 | nucleotide binding | IEA | - | |
| GO:0000287 | magnesium ion binding | IEA | - | |
| GO:0005515 | protein binding | IEA | - | |
| GO:0005524 | ATP binding | IEA | - | |
| GO:0004672 | protein kinase activity | IEA | - | |
| GO:0004674 | protein serine/threonine kinase activity | TAS | 8621389 | |
| GO:0004709 | MAP kinase kinase kinase activity | IEA | - | |
| GO:0016740 | transferase activity | IEA | - | |
| Biological process | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0006468 | protein amino acid phosphorylation | IEA | - | |
| GO:0007257 | activation of JNK activity | TAS | 8621389 | |
| Cellular component | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0005634 | nucleus | IEA | - | |
| GO:0005737 | cytoplasm | IEA | - |
Section IV. Protein-protein interaction annotation
| Interactors | Aliases B | Official full name B | Experimental | Source | PubMed ID |
|---|---|---|---|---|---|
| MAP2K4 | JNKK | JNKK1 | MAPKK4 | MEK4 | MKK4 | PRKMK4 | SEK1 | SERK1 | mitogen-activated protein kinase kinase 4 | - | HPRD | 7997270 |
| MAP2K5 | HsT17454 | MAPKK5 | MEK5 | PRKMK5 | mitogen-activated protein kinase kinase 5 | Affinity Capture-Western Two-hybrid | BioGRID | 11073940 |
| MAP2K5 | HsT17454 | MAPKK5 | MEK5 | PRKMK5 | mitogen-activated protein kinase kinase 5 | MEKK2 phosphorylates an unspecified isoform of MEK5. This interaction was modeled on a demonstrated interaction between MEKK2 from an unspecified species and MEK5 from an unspecified species. | BIND | 12659851 |
| MAP2K7 | Jnkk2 | MAPKK7 | MKK7 | PRKMK7 | mitogen-activated protein kinase kinase 7 | MEKK2 phosphorylates MKK7. This interaction was modeled on a demonstrated interaction between MEKK2 from an unspecified species and MKK7 from an unspecified species. | BIND | 12659851 |
| MAP2K7 | Jnkk2 | MAPKK7 | MKK7 | PRKMK7 | mitogen-activated protein kinase kinase 7 | - | HPRD,BioGRID | 10713157 |
| MAP3K2 | MEKK2 | MEKK2B | mitogen-activated protein kinase kinase kinase 2 | MEKK2 autophosphorylates. This interaction was modeled on a demonstrated interaction of MEKK2 from an unspecified species. | BIND | 12659851 |
| MAP3K5 | ASK1 | MAPKKK5 | MEKK5 | mitogen-activated protein kinase kinase kinase 5 | - | HPRD | 12912994 |
| MAPK8 | JNK | JNK1 | JNK1A2 | JNK21B1/2 | PRKM8 | SAPK1 | mitogen-activated protein kinase 8 | - | HPRD,BioGRID | 10713157 |
| PKN2 | MGC150606 | MGC71074 | PAK2 | PRK2 | PRKCL2 | PRO2042 | Pak-2 | protein kinase N2 | - | HPRD,BioGRID | 10818102 |
| SFN | YWHAS | stratifin | Affinity Capture-MS | BioGRID | 15778465 |
| SH2D2A | F2771 | TSAd | SH2 domain protein 2A | Affinity Capture-Western Two-hybrid | BioGRID | 11073940 |
| YWHAE | 14-3-3E | FLJ45465 | KCIP-1 | MDCR | MDS | tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein, epsilon polypeptide | - | HPRD,BioGRID | 9452471 |
| YWHAG | 14-3-3GAMMA | tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein, gamma polypeptide | - | HPRD | 15324660 |
| YWHAZ | KCIP-1 | MGC111427 | MGC126532 | MGC138156 | tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein, zeta polypeptide | - | HPRD | 9452471 |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| KEGG MAPK SIGNALING PATHWAY | 267 | 205 | All SZGR 2.0 genes in this pathway |
| KEGG GAP JUNCTION | 90 | 68 | All SZGR 2.0 genes in this pathway |
| KEGG GNRH SIGNALING PATHWAY | 101 | 77 | All SZGR 2.0 genes in this pathway |
| BIOCARTA MAPK PATHWAY | 87 | 68 | All SZGR 2.0 genes in this pathway |
| ST JNK MAPK PATHWAY | 40 | 30 | All SZGR 2.0 genes in this pathway |
| PID ERBB1 DOWNSTREAM PATHWAY | 105 | 78 | All SZGR 2.0 genes in this pathway |
| PID MAPK TRK PATHWAY | 34 | 31 | All SZGR 2.0 genes in this pathway |
| OSMAN BLADDER CANCER UP | 404 | 246 | All SZGR 2.0 genes in this pathway |
| OSWALD HEMATOPOIETIC STEM CELL IN COLLAGEN GEL DN | 275 | 168 | All SZGR 2.0 genes in this pathway |
| TONKS TARGETS OF RUNX1 RUNX1T1 FUSION MONOCYTE DN | 54 | 38 | All SZGR 2.0 genes in this pathway |
| SENESE HDAC1 TARGETS UP | 457 | 269 | All SZGR 2.0 genes in this pathway |
| SENESE HDAC3 TARGETS UP | 501 | 327 | All SZGR 2.0 genes in this pathway |
| HAMAI APOPTOSIS VIA TRAIL UP | 584 | 356 | All SZGR 2.0 genes in this pathway |
| NUYTTEN NIPP1 TARGETS DN | 848 | 527 | All SZGR 2.0 genes in this pathway |
| BUYTAERT PHOTODYNAMIC THERAPY STRESS UP | 811 | 508 | All SZGR 2.0 genes in this pathway |
| BENPORATH CYCLING GENES | 648 | 385 | All SZGR 2.0 genes in this pathway |
| STARK PREFRONTAL CORTEX 22Q11 DELETION UP | 195 | 138 | All SZGR 2.0 genes in this pathway |
| BYSTRYKH HEMATOPOIESIS STEM CELL QTL TRANS | 882 | 572 | All SZGR 2.0 genes in this pathway |
| CUI TCF21 TARGETS 2 DN | 830 | 547 | All SZGR 2.0 genes in this pathway |
| WHITFIELD CELL CYCLE S | 162 | 86 | All SZGR 2.0 genes in this pathway |
| STAMBOLSKY TARGETS OF MUTATED TP53 UP | 49 | 35 | All SZGR 2.0 genes in this pathway |
| GABRIELY MIR21 TARGETS | 289 | 187 | All SZGR 2.0 genes in this pathway |
| JOHNSTONE PARVB TARGETS 3 DN | 918 | 550 | All SZGR 2.0 genes in this pathway |
| LINSLEY MIR16 TARGETS | 206 | 127 | All SZGR 2.0 genes in this pathway |
| PLASARI TGFB1 SIGNALING VIA NFIC 1HR DN | 106 | 77 | All SZGR 2.0 genes in this pathway |
| FORTSCHEGGER PHF8 TARGETS DN | 784 | 464 | All SZGR 2.0 genes in this pathway |
| RATTENBACHER BOUND BY CELF1 | 467 | 251 | All SZGR 2.0 genes in this pathway |
| ZWANG TRANSIENTLY UP BY 1ST EGF PULSE ONLY | 1839 | 928 | All SZGR 2.0 genes in this pathway |
Section VI. microRNA annotation
| miRNA family | Target position | miRNA ID | miRNA seq | ||
|---|---|---|---|---|---|
| UTR start | UTR end | Match method | |||
| miR-10 | 2407 | 2413 | 1A | hsa-miR-10a | UACCCUGUAGAUCCGAAUUUGUG |
| hsa-miR-10b | UACCCUGUAGAACCGAAUUUGU | ||||
| miR-122 | 2868 | 2874 | m8 | hsa-miR-122a | UGGAGUGUGACAAUGGUGUUUGU |
| miR-129-5p | 2763 | 2769 | 1A | hsa-miR-129brain | CUUUUUGCGGUCUGGGCUUGC |
| hsa-miR-129-5p | CUUUUUGCGGUCUGGGCUUGCU | ||||
| miR-17-5p/20/93.mr/106/519.d | 65 | 71 | m8 | hsa-miR-17-5p | CAAAGUGCUUACAGUGCAGGUAGU |
| hsa-miR-20abrain | UAAAGUGCUUAUAGUGCAGGUAG | ||||
| hsa-miR-106a | AAAAGUGCUUACAGUGCAGGUAGC | ||||
| hsa-miR-106bSZ | UAAAGUGCUGACAGUGCAGAU | ||||
| hsa-miR-20bSZ | CAAAGUGCUCAUAGUGCAGGUAG | ||||
| hsa-miR-519d | CAAAGUGCCUCCCUUUAGAGUGU | ||||
| hsa-miR-17-5p | CAAAGUGCUUACAGUGCAGGUAGU | ||||
| hsa-miR-20abrain | UAAAGUGCUUAUAGUGCAGGUAG | ||||
| hsa-miR-106a | AAAAGUGCUUACAGUGCAGGUAGC | ||||
| hsa-miR-106bSZ | UAAAGUGCUGACAGUGCAGAU | ||||
| hsa-miR-20bSZ | CAAAGUGCUCAUAGUGCAGGUAG | ||||
| hsa-miR-519d | CAAAGUGCCUCCCUUUAGAGUGU | ||||
| miR-186 | 522 | 529 | 1A,m8 | hsa-miR-186 | CAAAGAAUUCUCCUUUUGGGCUU |
| miR-223 | 346 | 352 | 1A | hsa-miR-223 | UGUCAGUUUGUCAAAUACCCC |
| miR-23 | 2686 | 2692 | 1A | hsa-miR-23abrain | AUCACAUUGCCAGGGAUUUCC |
| hsa-miR-23bbrain | AUCACAUUGCCAGGGAUUACC | ||||
| miR-26 | 208 | 214 | 1A | hsa-miR-26abrain | UUCAAGUAAUCCAGGAUAGGC |
| hsa-miR-26bSZ | UUCAAGUAAUUCAGGAUAGGUU | ||||
| miR-335 | 128 | 134 | 1A | hsa-miR-335brain | UCAAGAGCAAUAACGAAAAAUGU |
| miR-539 | 124 | 130 | m8 | hsa-miR-539 | GGAGAAAUUAUCCUUGGUGUGU |
- SZ: miRNAs which differentially expressed in brain cortex of schizophrenia patients comparing with control samples using microarray. Click here to see the list of SZ related miRNAs.
- Brain: miRNAs which are expressed in brain based on miRNA microarray expression studies. Click here to see the list of brain related miRNAs.