Gene Page: TCERG1
Summary ?
| GeneID | 10915 |
| Symbol | TCERG1 |
| Synonyms | CA150|TAF2S|Urn1 |
| Description | transcription elongation regulator 1 |
| Reference | MIM:605409|HGNC:HGNC:15630|Ensembl:ENSG00000113649|HPRD:10393|Vega:OTTHUMG00000129683 |
| Gene type | protein-coding |
| Map location | 5q31 |
| Pascal p-value | 0.599 |
| Sherlock p-value | 0.376 |
| DEG p-value | DEG:Zhao_2015:p=6.61e-04:q=0.0955 |
| Fetal beta | 1.105 |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CV:GWASdb | Genome-wide Association Studies | GWASdb records for schizophrenia | |
| CV:PGCnp | Genome-wide Association Study | GWAS | |
| DEG:Zhao_2015 | RNA Sequencing analysis | Transcriptome sequencing and genome-wide association analyses reveal lysosomal function and actin cytoskeleton remodeling in schizophrenia and bipolar disorder. | |
| GSMA_I | Genome scan meta-analysis | Psr: 0.0032 | |
| GSMA_IIE | Genome scan meta-analysis (European-ancestry samples) | Psr: 0.01718 | |
| GSMA_IIA | Genome scan meta-analysis (All samples) | Psr: 0.00459 |
Section I. Genetics and epigenetics annotation
Section II. Transcriptome annotation
General gene expression (GTEx)
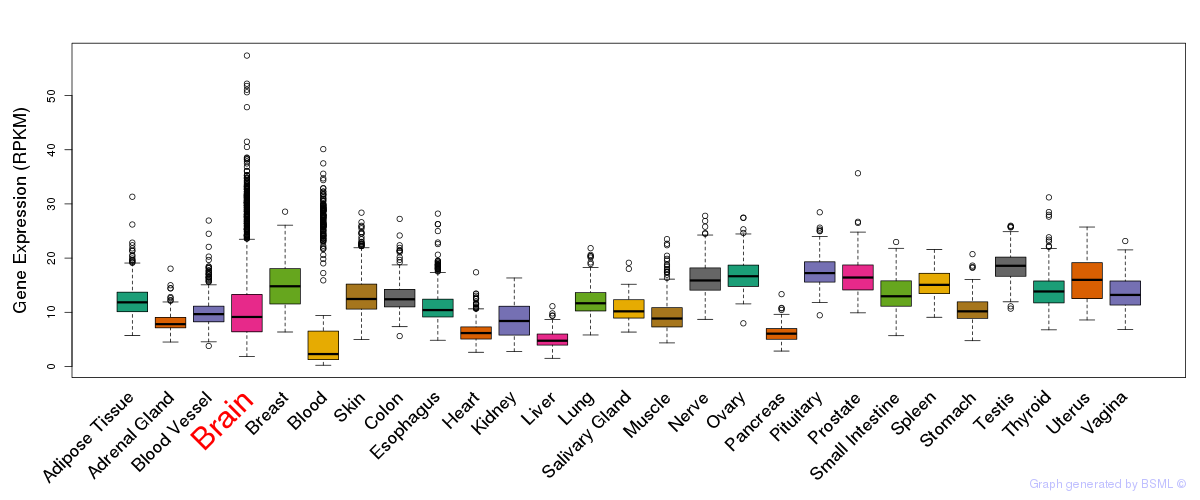
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
Top co-expressed genes in brain regions
| Top 10 positively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| CKAP5 | 0.96 | 0.97 |
| RBM16 | 0.96 | 0.97 |
| SMEK1 | 0.95 | 0.96 |
| KIAA0947 | 0.95 | 0.96 |
| NFX1 | 0.95 | 0.96 |
| NUP153 | 0.94 | 0.96 |
| HP1BP3 | 0.94 | 0.96 |
| FBXO11 | 0.94 | 0.96 |
| RNASEN | 0.94 | 0.95 |
| TTC3 | 0.94 | 0.97 |
| Top 10 negatively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| AF347015.31 | -0.82 | -0.90 |
| MT-CO2 | -0.82 | -0.91 |
| FXYD1 | -0.81 | -0.89 |
| AF347015.27 | -0.80 | -0.87 |
| AF347015.33 | -0.80 | -0.87 |
| MT-CYB | -0.79 | -0.87 |
| AF347015.8 | -0.78 | -0.89 |
| HIGD1B | -0.78 | -0.89 |
| S100B | -0.78 | -0.84 |
| IFI27 | -0.78 | -0.87 |
Section III. Gene Ontology annotation
| Molecular function | GO term | Evidence | Neuro keywords | PubMed ID |
|---|---|---|---|---|
| GO:0003702 | RNA polymerase II transcription factor activity | TAS | 9315662 | |
| GO:0003713 | transcription coactivator activity | TAS | 9315662 | |
| GO:0005515 | protein binding | IPI | 15383276 | |
| Biological process | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0006355 | regulation of transcription, DNA-dependent | IEA | - | |
| GO:0006350 | transcription | IEA | - | |
| GO:0006366 | transcription from RNA polymerase II promoter | TAS | 9315662 | |
| Cellular component | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0005634 | nucleus | TAS | 9315662 |
Section IV. Protein-protein interaction annotation
| Interactors | Aliases B | Official full name B | Experimental | Source | PubMed ID |
|---|---|---|---|---|---|
| BARD1 | - | BRCA1 associated RING domain 1 | - | HPRD | 15383276 |
| CHAF1A | CAF-1 | CAF1 | CAF1B | CAF1P150 | MGC71229 | P150 | chromatin assembly factor 1, subunit A (p150) | - | HPRD,BioGRID | 15456888 |
| CHD8 | DKFZp686N17164 | HELSNF1 | KIAA1564 | chromodomain helicase DNA binding protein 8 | Affinity Capture-MS | BioGRID | 15456888 |
| DNAJB9 | DKFZp564F1862 | ERdj4 | MDG1 | MST049 | MSTP049 | DnaJ (Hsp40) homolog, subfamily B, member 9 | - | HPRD,BioGRID | 15456888 |
| FANCA | FA | FA-H | FA1 | FAA | FACA | FAH | FANCH | MGC75158 | Fanconi anemia, complementation group A | Two-hybrid | BioGRID | 14499622 |
| FANCC | FA3 | FAC | FACC | FLJ14675 | Fanconi anemia, complementation group C | Two-hybrid | BioGRID | 14499622 |
| HNRNPH1 | DKFZp686A15170 | HNRPH | HNRPH1 | hnRNPH | heterogeneous nuclear ribonucleoprotein H1 (H) | - | HPRD,BioGRID | 15456888 |
| HNRNPM | DKFZp547H118 | HNRNPM4 | HNRPM | HNRPM4 | HTGR1 | NAGR1 | heterogeneous nuclear ribonucleoprotein M | - | HPRD,BioGRID | 15456888 |
| HSPA1A | FLJ54303 | FLJ54370 | FLJ54392 | FLJ54408 | FLJ75127 | HSP70-1 | HSP70-1A | HSP70I | HSP72 | HSPA1 | HSPA1B | heat shock 70kDa protein 1A | - | HPRD | 15456888 |
| HSPA4 | APG-2 | HS24/P52 | MGC131852 | RY | hsp70 | hsp70RY | heat shock 70kDa protein 4 | Affinity Capture-MS | BioGRID | 15456888 |
| HSPA5 | BIP | FLJ26106 | GRP78 | MIF2 | heat shock 70kDa protein 5 (glucose-regulated protein, 78kDa) | - | HPRD,BioGRID | 15456888 |
| HSPA8 | HSC54 | HSC70 | HSC71 | HSP71 | HSP73 | HSPA10 | LAP1 | MGC131511 | MGC29929 | NIP71 | heat shock 70kDa protein 8 | - | HPRD,BioGRID | 15456888 |
| HTT | HD | IT15 | huntingtin | Htt interacts with CA150. | BIND | 11172033 |
| HTT | HD | IT15 | huntingtin | - | HPRD,BioGRID | 11172033 |
| LUC7L | FLJ10231 | LUC7-LIKE | LUC7B1 | SR+89 | LUC7-like (S. cerevisiae) | - | HPRD | 15383276 |
| MED21 | SRB7 | SURB7 | mediator complex subunit 21 | Affinity Capture-Western | BioGRID | 9315662 |
| NPM1 | B23 | MGC104254 | NPM | nucleophosmin (nucleolar phosphoprotein B23, numatrin) | - | HPRD,BioGRID | 15456888 |
| PCBP1 | HNRPE1 | HNRPX | hnRNP-E1 | hnRNP-X | poly(rC) binding protein 1 | - | HPRD,BioGRID | 15456888 |
| PIAS4 | FLJ12419 | MGC35296 | PIASY | Piasg | ZMIZ6 | protein inhibitor of activated STAT, 4 | - | HPRD | 15383276 |
| POLR2A | MGC75453 | POLR2 | POLRA | RPB1 | RPBh1 | RPO2 | RPOL2 | RpIILS | hRPB220 | hsRPB1 | polymerase (RNA) II (DNA directed) polypeptide A, 220kDa | - | HPRD,BioGRID | 9315662 |10908677 |
| RNU1A | HU1-1 | RNU1 | RNU1G4 | Rnu1a1 | U1 | RNA, U1A small nuclear | Affinity Capture-RNA | BioGRID | 15456888 |
| RNU2-1 | RNU2 | U2 | RNA, U2 small nuclear 1 | Affinity Capture-RNA | BioGRID | 15456888 |
| SETDB1 | ESET | KG1T | KIAA0067 | KMT1E | SET domain, bifurcated 1 | - | HPRD | 15383276 |
| SF1 | D11S636 | ZFM1 | ZNF162 | splicing factor 1 | - | HPRD,BioGRID | 11604498 |
| SF3A1 | PRP21 | PRPF21 | SAP114 | SF3A120 | splicing factor 3a, subunit 1, 120kDa | - | HPRD,BioGRID | 15456888 |
| SF3A3 | PRP9 | PRPF9 | SAP61 | SF3a60 | splicing factor 3a, subunit 3, 60kDa | - | HPRD,BioGRID | 15456888 |
| SF3B1 | PRP10 | PRPF10 | SAP155 | SF3b155 | splicing factor 3b, subunit 1, 155kDa | - | HPRD,BioGRID | 15456888 |
| SF3B2 | SAP145 | SF3B145 | SF3b1 | SF3b150 | splicing factor 3b, subunit 2, 145kDa | - | HPRD,BioGRID | 15456888 |
| SF3B4 | MGC10828 | SAP49 | SF3b49 | splicing factor 3b, subunit 4, 49kDa | - | HPRD,BioGRID | 15456888 |
| SFRS1 | ASF | MGC5228 | SF2 | SF2p33 | SRp30a | splicing factor, arginine/serine-rich 1 | - | HPRD,BioGRID | 15456888 |
| TPM4 | - | tropomyosin 4 | - | HPRD,BioGRID | 15456888 |
| U2AF2 | U2AF65 | U2 small nuclear RNA auxiliary factor 2 | - | HPRD,BioGRID | 15456888 |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| KEGG SPLICEOSOME | 128 | 72 | All SZGR 2.0 genes in this pathway |
| ONKEN UVEAL MELANOMA UP | 783 | 507 | All SZGR 2.0 genes in this pathway |
| DIAZ CHRONIC MEYLOGENOUS LEUKEMIA UP | 1382 | 904 | All SZGR 2.0 genes in this pathway |
| TAKEDA TARGETS OF NUP98 HOXA9 FUSION 8D DN | 205 | 127 | All SZGR 2.0 genes in this pathway |
| ENK UV RESPONSE KERATINOCYTE DN | 485 | 334 | All SZGR 2.0 genes in this pathway |
| GRAESSMANN APOPTOSIS BY DOXORUBICIN DN | 1781 | 1082 | All SZGR 2.0 genes in this pathway |
| GRAESSMANN RESPONSE TO MC AND DOXORUBICIN DN | 770 | 415 | All SZGR 2.0 genes in this pathway |
| BERENJENO TRANSFORMED BY RHOA UP | 536 | 340 | All SZGR 2.0 genes in this pathway |
| SEIDEN ONCOGENESIS BY MET | 88 | 53 | All SZGR 2.0 genes in this pathway |
| SCHLOSSER MYC TARGETS REPRESSED BY SERUM | 159 | 93 | All SZGR 2.0 genes in this pathway |
| PUJANA XPRSS INT NETWORK | 168 | 103 | All SZGR 2.0 genes in this pathway |
| PUJANA BRCA1 PCC NETWORK | 1652 | 1023 | All SZGR 2.0 genes in this pathway |
| PUJANA BRCA2 PCC NETWORK | 423 | 265 | All SZGR 2.0 genes in this pathway |
| PUJANA ATM PCC NETWORK | 1442 | 892 | All SZGR 2.0 genes in this pathway |
| PUJANA CHEK2 PCC NETWORK | 779 | 480 | All SZGR 2.0 genes in this pathway |
| PUJANA BRCA CENTERED NETWORK | 117 | 72 | All SZGR 2.0 genes in this pathway |
| NUYTTEN EZH2 TARGETS DN | 1024 | 594 | All SZGR 2.0 genes in this pathway |
| MANTOVANI VIRAL GPCR SIGNALING DN | 49 | 37 | All SZGR 2.0 genes in this pathway |
| BENPORATH NANOG TARGETS | 988 | 594 | All SZGR 2.0 genes in this pathway |
| LI WILMS TUMOR VS FETAL KIDNEY 1 DN | 163 | 115 | All SZGR 2.0 genes in this pathway |
| BLALOCK ALZHEIMERS DISEASE DN | 1237 | 837 | All SZGR 2.0 genes in this pathway |
| WANG SMARCE1 TARGETS DN | 371 | 218 | All SZGR 2.0 genes in this pathway |
| YAMAZAKI TCEB3 TARGETS UP | 175 | 116 | All SZGR 2.0 genes in this pathway |
| MARTINEZ RESPONSE TO TRABECTEDIN DN | 271 | 175 | All SZGR 2.0 genes in this pathway |
| MARSON BOUND BY E2F4 UNSTIMULATED | 728 | 415 | All SZGR 2.0 genes in this pathway |
| MUELLER PLURINET | 299 | 189 | All SZGR 2.0 genes in this pathway |
| SHAFFER IRF4 TARGETS IN ACTIVATED DENDRITIC CELL | 65 | 49 | All SZGR 2.0 genes in this pathway |
| PUJANA BREAST CANCER WITH BRCA1 MUTATED UP | 56 | 27 | All SZGR 2.0 genes in this pathway |
| CAIRO HEPATOBLASTOMA CLASSES UP | 605 | 377 | All SZGR 2.0 genes in this pathway |
| YAO TEMPORAL RESPONSE TO PROGESTERONE CLUSTER 14 | 143 | 86 | All SZGR 2.0 genes in this pathway |
| WHITFIELD CELL CYCLE M G1 | 148 | 95 | All SZGR 2.0 genes in this pathway |
| PILON KLF1 TARGETS DN | 1972 | 1213 | All SZGR 2.0 genes in this pathway |
| JOHNSTONE PARVB TARGETS 2 DN | 336 | 211 | All SZGR 2.0 genes in this pathway |
| LEE BMP2 TARGETS DN | 882 | 538 | All SZGR 2.0 genes in this pathway |
Section VI. microRNA annotation
| miRNA family | Target position | miRNA ID | miRNA seq | ||
|---|---|---|---|---|---|
| UTR start | UTR end | Match method | |||
| miR-141/200a | 300 | 307 | 1A,m8 | hsa-miR-141 | UAACACUGUCUGGUAAAGAUGG |
| hsa-miR-200a | UAACACUGUCUGGUAACGAUGU | ||||
| miR-181 | 773 | 779 | m8 | hsa-miR-181abrain | AACAUUCAACGCUGUCGGUGAGU |
| hsa-miR-181bSZ | AACAUUCAUUGCUGUCGGUGGG | ||||
| hsa-miR-181cbrain | AACAUUCAACCUGUCGGUGAGU | ||||
| hsa-miR-181dbrain | AACAUUCAUUGUUGUCGGUGGGUU | ||||
| hsa-miR-181abrain | AACAUUCAACGCUGUCGGUGAGU | ||||
| hsa-miR-181bSZ | AACAUUCAUUGCUGUCGGUGGG | ||||
| hsa-miR-181cbrain | AACAUUCAACCUGUCGGUGAGU | ||||
| hsa-miR-181dbrain | AACAUUCAUUGUUGUCGGUGGGUU | ||||
| miR-223 | 187 | 193 | 1A | hsa-miR-223 | UGUCAGUUUGUCAAAUACCCC |
| miR-224 | 276 | 282 | m8 | hsa-miR-224 | CAAGUCACUAGUGGUUCCGUUUA |
| miR-24 | 45 | 51 | 1A | hsa-miR-24SZ | UGGCUCAGUUCAGCAGGAACAG |
| miR-299-5p | 846 | 852 | 1A | hsa-miR-299-5p | UGGUUUACCGUCCCACAUACAU |
| miR-369-3p | 475 | 481 | 1A | hsa-miR-369-3p | AAUAAUACAUGGUUGAUCUUU |
| miR-374 | 475 | 482 | 1A,m8 | hsa-miR-374 | UUAUAAUACAACCUGAUAAGUG |
| miR-543 | 816 | 822 | m8 | hsa-miR-543 | AAACAUUCGCGGUGCACUUCU |
| hsa-miR-543 | AAACAUUCGCGGUGCACUUCU | ||||
| miR-7 | 457 | 463 | 1A | hsa-miR-7SZ | UGGAAGACUAGUGAUUUUGUUG |
- SZ: miRNAs which differentially expressed in brain cortex of schizophrenia patients comparing with control samples using microarray. Click here to see the list of SZ related miRNAs.
- Brain: miRNAs which are expressed in brain based on miRNA microarray expression studies. Click here to see the list of brain related miRNAs.