Gene Page: SDS
Summary ?
| GeneID | 10993 |
| Symbol | SDS |
| Synonyms | SDH |
| Description | serine dehydratase |
| Reference | MIM:182128|HGNC:HGNC:10691|Ensembl:ENSG00000135094|HPRD:01633|Vega:OTTHUMG00000169554 |
| Gene type | protein-coding |
| Map location | 12q24.13 |
| Pascal p-value | 9.755E-5 |
| Fetal beta | -1.765 |
| eGene | Myers' cis & trans |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CV:PGCnp | Genome-wide Association Study | GWAS | |
| PMID:cooccur | High-throughput literature-search | Systematic search in PubMed for genes co-occurring with SCZ keywords. A total of 3027 genes were included. | |
| Literature | High-throughput literature-search | Co-occurance with Schizophrenia keywords: schizophrenia,schizophrenic,schizophrenics,schizotypy,schizophrenias,schizotypal | Click to show details |
Section I. Genetics and epigenetics annotation
 eQTL annotation
eQTL annotation
| SNP ID | Chromosome | Position | eGene | Gene Entrez ID | pvalue | qvalue | TSS distance | eQTL type |
|---|---|---|---|---|---|---|---|---|
| rs17455108 | chr1 | 4724047 | SDS | 10993 | 0.01 | trans | ||
| rs6703905 | chr1 | 89656748 | SDS | 10993 | 0.01 | trans | ||
| rs924019 | chr3 | 184794255 | SDS | 10993 | 0.07 | trans | ||
| rs17007771 | chr4 | 124970238 | SDS | 10993 | 0.01 | trans | ||
| rs17007781 | chr4 | 124977313 | SDS | 10993 | 0.03 | trans | ||
| rs1028198 | chr4 | 124977733 | SDS | 10993 | 0.02 | trans | ||
| rs226946 | chr6 | 3717139 | SDS | 10993 | 0.19 | trans | ||
| rs226966 | chr6 | 3720300 | SDS | 10993 | 0.14 | trans | ||
| rs3803656 | chr16 | 81551505 | SDS | 10993 | 0 | trans |
Section II. Transcriptome annotation
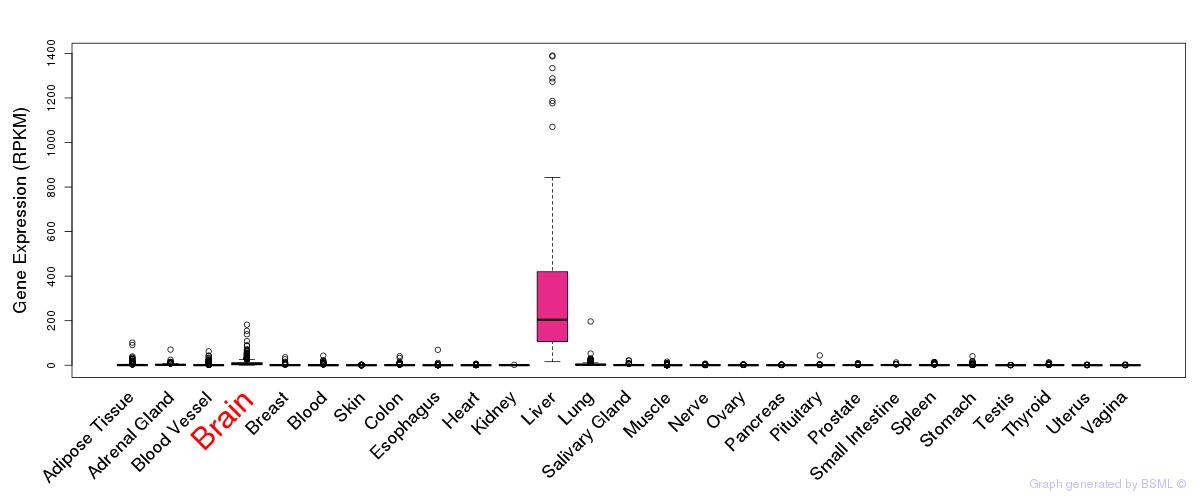
General gene expression (GTEx)
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
Top co-expressed genes in brain regions
| Top 10 positively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| SLC25A11 | 0.90 | 0.90 |
| MRPL38 | 0.89 | 0.91 |
| TMEM222 | 0.88 | 0.88 |
| UQCRC1 | 0.88 | 0.87 |
| MRPS18B | 0.88 | 0.84 |
| AES | 0.88 | 0.88 |
| PSMD8 | 0.88 | 0.84 |
| C21orf33 | 0.87 | 0.85 |
| NIT1 | 0.86 | 0.83 |
| FBXW5 | 0.86 | 0.87 |
| Top 10 negatively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| AF347015.18 | -0.60 | -0.49 |
| MT-ATP8 | -0.55 | -0.42 |
| AL139819.3 | -0.55 | -0.49 |
| AC010300.1 | -0.54 | -0.59 |
| NSBP1 | -0.54 | -0.53 |
| AF347015.21 | -0.53 | -0.38 |
| AF347015.8 | -0.53 | -0.37 |
| AF347015.26 | -0.52 | -0.38 |
| AF347015.2 | -0.52 | -0.34 |
| MT-CYB | -0.49 | -0.34 |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| KEGG GLYCINE SERINE AND THREONINE METABOLISM | 31 | 26 | All SZGR 2.0 genes in this pathway |
| KEGG CYSTEINE AND METHIONINE METABOLISM | 34 | 24 | All SZGR 2.0 genes in this pathway |
| FULCHER INFLAMMATORY RESPONSE LECTIN VS LPS DN | 463 | 290 | All SZGR 2.0 genes in this pathway |
| TAKEDA TARGETS OF NUP98 HOXA9 FUSION 16D DN | 143 | 83 | All SZGR 2.0 genes in this pathway |
| MULLIGHAN NPM1 MUTATED SIGNATURE 1 UP | 276 | 165 | All SZGR 2.0 genes in this pathway |
| MULLIGHAN NPM1 SIGNATURE 3 UP | 341 | 197 | All SZGR 2.0 genes in this pathway |
| HSIAO LIVER SPECIFIC GENES | 244 | 154 | All SZGR 2.0 genes in this pathway |
| WENG POR TARGETS LIVER DN | 20 | 11 | All SZGR 2.0 genes in this pathway |
| MOOTHA HUMAN MITODB 6 2002 | 429 | 260 | All SZGR 2.0 genes in this pathway |
| YOSHIMURA MAPK8 TARGETS UP | 1305 | 895 | All SZGR 2.0 genes in this pathway |
| HOSHIDA LIVER CANCER SUBCLASS S3 | 266 | 180 | All SZGR 2.0 genes in this pathway |
| CAIRO HEPATOBLASTOMA DN | 267 | 160 | All SZGR 2.0 genes in this pathway |
| BOSCO TH1 CYTOTOXIC MODULE | 114 | 62 | All SZGR 2.0 genes in this pathway |