| KEGG ENDOCYTOSIS
| 183 | 132 | All SZGR 2.0 genes in this pathway |
| NAKAMURA TUMOR ZONE PERIPHERAL VS CENTRAL DN
| 634 | 384 | All SZGR 2.0 genes in this pathway |
| LIU SOX4 TARGETS DN
| 309 | 191 | All SZGR 2.0 genes in this pathway |
| ONKEN UVEAL MELANOMA UP
| 783 | 507 | All SZGR 2.0 genes in this pathway |
| FULCHER INFLAMMATORY RESPONSE LECTIN VS LPS UP
| 579 | 346 | All SZGR 2.0 genes in this pathway |
| DIAZ CHRONIC MEYLOGENOUS LEUKEMIA UP
| 1382 | 904 | All SZGR 2.0 genes in this pathway |
| CHARAFE BREAST CANCER LUMINAL VS MESENCHYMAL DN
| 460 | 312 | All SZGR 2.0 genes in this pathway |
| ROY WOUND BLOOD VESSEL UP
| 50 | 30 | All SZGR 2.0 genes in this pathway |
| VECCHI GASTRIC CANCER ADVANCED VS EARLY UP
| 175 | 120 | All SZGR 2.0 genes in this pathway |
| HOEBEKE LYMPHOID STEM CELL UP
| 95 | 64 | All SZGR 2.0 genes in this pathway |
| GRAHAM CML DIVIDING VS NORMAL QUIESCENT UP
| 181 | 101 | All SZGR 2.0 genes in this pathway |
| ENK UV RESPONSE EPIDERMIS UP
| 293 | 179 | All SZGR 2.0 genes in this pathway |
| HERNANDEZ ABERRANT MITOSIS BY DOCETACEL 2NM UP
| 81 | 57 | All SZGR 2.0 genes in this pathway |
| KANG GIST WITH PDGFRA UP
| 50 | 27 | All SZGR 2.0 genes in this pathway |
| DACOSTA UV RESPONSE VIA ERCC3 DN
| 855 | 609 | All SZGR 2.0 genes in this pathway |
| GRUETZMANN PANCREATIC CANCER UP
| 358 | 245 | All SZGR 2.0 genes in this pathway |
| NUYTTEN EZH2 TARGETS DN
| 1024 | 594 | All SZGR 2.0 genes in this pathway |
| LOPEZ MBD TARGETS
| 957 | 597 | All SZGR 2.0 genes in this pathway |
| RICKMAN TUMOR DIFFERENTIATED WELL VS POORLY DN
| 382 | 224 | All SZGR 2.0 genes in this pathway |
| RICKMAN TUMOR DIFFERENTIATED WELL VS MODERATELY DN
| 110 | 64 | All SZGR 2.0 genes in this pathway |
| FRIDMAN SENESCENCE UP
| 77 | 60 | All SZGR 2.0 genes in this pathway |
| WU CELL MIGRATION
| 184 | 114 | All SZGR 2.0 genes in this pathway |
| BENPORATH SUZ12 TARGETS
| 1038 | 678 | All SZGR 2.0 genes in this pathway |
| LAMB CCND1 TARGETS
| 19 | 14 | All SZGR 2.0 genes in this pathway |
| FRASOR RESPONSE TO ESTRADIOL UP
| 37 | 28 | All SZGR 2.0 genes in this pathway |
| DORSAM HOXA9 TARGETS UP
| 35 | 25 | All SZGR 2.0 genes in this pathway |
| FLECHNER BIOPSY KIDNEY TRANSPLANT REJECTED VS OK UP
| 87 | 58 | All SZGR 2.0 genes in this pathway |
| WIELAND UP BY HBV INFECTION
| 101 | 66 | All SZGR 2.0 genes in this pathway |
| OKUMURA INFLAMMATORY RESPONSE LPS
| 183 | 115 | All SZGR 2.0 genes in this pathway |
| PENG GLUCOSE DEPRIVATION DN
| 169 | 112 | All SZGR 2.0 genes in this pathway |
| ZHAN MULTIPLE MYELOMA CD1 UP
| 45 | 29 | All SZGR 2.0 genes in this pathway |
| FAELT B CLL WITH VH REARRANGEMENTS UP
| 48 | 34 | All SZGR 2.0 genes in this pathway |
| ZHAN MULTIPLE MYELOMA CD1 VS CD2 UP
| 66 | 47 | All SZGR 2.0 genes in this pathway |
| PETROVA ENDOTHELIUM LYMPHATIC VS BLOOD UP
| 131 | 87 | All SZGR 2.0 genes in this pathway |
| HADDAD T LYMPHOCYTE AND NK PROGENITOR DN
| 63 | 41 | All SZGR 2.0 genes in this pathway |
| CROMER METASTASIS DN
| 81 | 58 | All SZGR 2.0 genes in this pathway |
| TAVOR CEBPA TARGETS UP
| 48 | 36 | All SZGR 2.0 genes in this pathway |
| FAELT B CLL WITH VH3 21 DN
| 49 | 29 | All SZGR 2.0 genes in this pathway |
| KUMAR TARGETS OF MLL AF9 FUSION
| 405 | 264 | All SZGR 2.0 genes in this pathway |
| REN ALVEOLAR RHABDOMYOSARCOMA DN
| 408 | 274 | All SZGR 2.0 genes in this pathway |
| IVANOVA HEMATOPOIESIS LATE PROGENITOR
| 544 | 307 | All SZGR 2.0 genes in this pathway |
| BLALOCK ALZHEIMERS DISEASE UP
| 1691 | 1088 | All SZGR 2.0 genes in this pathway |
| BURTON ADIPOGENESIS 8
| 86 | 57 | All SZGR 2.0 genes in this pathway |
| MCLACHLAN DENTAL CARIES DN
| 245 | 144 | All SZGR 2.0 genes in this pathway |
| RODWELL AGING KIDNEY UP
| 487 | 303 | All SZGR 2.0 genes in this pathway |
| XU GH1 AUTOCRINE TARGETS DN
| 142 | 94 | All SZGR 2.0 genes in this pathway |
| SATO SILENCED BY METHYLATION IN PANCREATIC CANCER 1
| 419 | 273 | All SZGR 2.0 genes in this pathway |
| MCLACHLAN DENTAL CARIES UP
| 253 | 147 | All SZGR 2.0 genes in this pathway |
| CREIGHTON ENDOCRINE THERAPY RESISTANCE 1
| 528 | 324 | All SZGR 2.0 genes in this pathway |
| CREIGHTON ENDOCRINE THERAPY RESISTANCE 4
| 307 | 185 | All SZGR 2.0 genes in this pathway |
| RIGGINS TAMOXIFEN RESISTANCE DN
| 220 | 147 | All SZGR 2.0 genes in this pathway |
| STEIN ESRRA TARGETS RESPONSIVE TO ESTROGEN DN
| 41 | 26 | All SZGR 2.0 genes in this pathway |
| MARTINEZ RB1 TARGETS UP
| 673 | 430 | All SZGR 2.0 genes in this pathway |
| MARTINEZ TP53 TARGETS UP
| 602 | 364 | All SZGR 2.0 genes in this pathway |
| MARTINEZ RB1 AND TP53 TARGETS UP
| 601 | 369 | All SZGR 2.0 genes in this pathway |
| MASSARWEH TAMOXIFEN RESISTANCE DN
| 258 | 160 | All SZGR 2.0 genes in this pathway |
| TOYOTA TARGETS OF MIR34B AND MIR34C
| 463 | 262 | All SZGR 2.0 genes in this pathway |
| IZADPANAH STEM CELL ADIPOSE VS BONE UP
| 126 | 92 | All SZGR 2.0 genes in this pathway |
| MOOTHA PGC
| 420 | 269 | All SZGR 2.0 genes in this pathway |
| LINDSTEDT DENDRITIC CELL MATURATION D
| 68 | 44 | All SZGR 2.0 genes in this pathway |
| PARK APL PATHOGENESIS DN
| 50 | 35 | All SZGR 2.0 genes in this pathway |
| CHEN METABOLIC SYNDROM NETWORK
| 1210 | 725 | All SZGR 2.0 genes in this pathway |
| BOYAULT LIVER CANCER SUBCLASS G5 DN
| 27 | 17 | All SZGR 2.0 genes in this pathway |
| STEIN ESR1 TARGETS
| 85 | 55 | All SZGR 2.0 genes in this pathway |
| HOSHIDA LIVER CANCER SUBCLASS S1
| 237 | 159 | All SZGR 2.0 genes in this pathway |
| NAKAMURA ADIPOGENESIS LATE UP
| 104 | 67 | All SZGR 2.0 genes in this pathway |
| SASSON RESPONSE TO GONADOTROPHINS UP
| 91 | 59 | All SZGR 2.0 genes in this pathway |
| SASSON RESPONSE TO FORSKOLIN DN
| 88 | 68 | All SZGR 2.0 genes in this pathway |
| RAGHAVACHARI PLATELET SPECIFIC GENES
| 70 | 46 | All SZGR 2.0 genes in this pathway |
| KAMIKUBO MYELOID CEBPA NETWORK
| 28 | 19 | All SZGR 2.0 genes in this pathway |
| MARTENS BOUND BY PML RARA FUSION
| 456 | 287 | All SZGR 2.0 genes in this pathway |
| CHICAS RB1 TARGETS SENESCENT
| 572 | 352 | All SZGR 2.0 genes in this pathway |
| HOELZEL NF1 TARGETS UP
| 139 | 93 | All SZGR 2.0 genes in this pathway |
| DUTERTRE ESTRADIOL RESPONSE 6HR UP
| 229 | 149 | All SZGR 2.0 genes in this pathway |
| DUTERTRE ESTRADIOL RESPONSE 24HR UP
| 324 | 193 | All SZGR 2.0 genes in this pathway |
| CHYLA CBFA2T3 TARGETS UP
| 387 | 225 | All SZGR 2.0 genes in this pathway |
| PILON KLF1 TARGETS DN
| 1972 | 1213 | All SZGR 2.0 genes in this pathway |
| BRUINS UVC RESPONSE VIA TP53 GROUP A
| 898 | 516 | All SZGR 2.0 genes in this pathway |
| BRUINS UVC RESPONSE LATE
| 1137 | 655 | All SZGR 2.0 genes in this pathway |
| LEE BMP2 TARGETS UP
| 745 | 475 | All SZGR 2.0 genes in this pathway |
| HUANG GATA2 TARGETS UP
| 149 | 96 | All SZGR 2.0 genes in this pathway |
| FORTSCHEGGER PHF8 TARGETS DN
| 784 | 464 | All SZGR 2.0 genes in this pathway |
| BOUDOUKHA BOUND BY IGF2BP2
| 111 | 59 | All SZGR 2.0 genes in this pathway |
| ACEVEDO FGFR1 TARGETS IN PROSTATE CANCER MODEL UP
| 289 | 184 | All SZGR 2.0 genes in this pathway |
| ANASTASSIOU CANCER MESENCHYMAL TRANSITION SIGNATURE
| 64 | 40 | All SZGR 2.0 genes in this pathway |
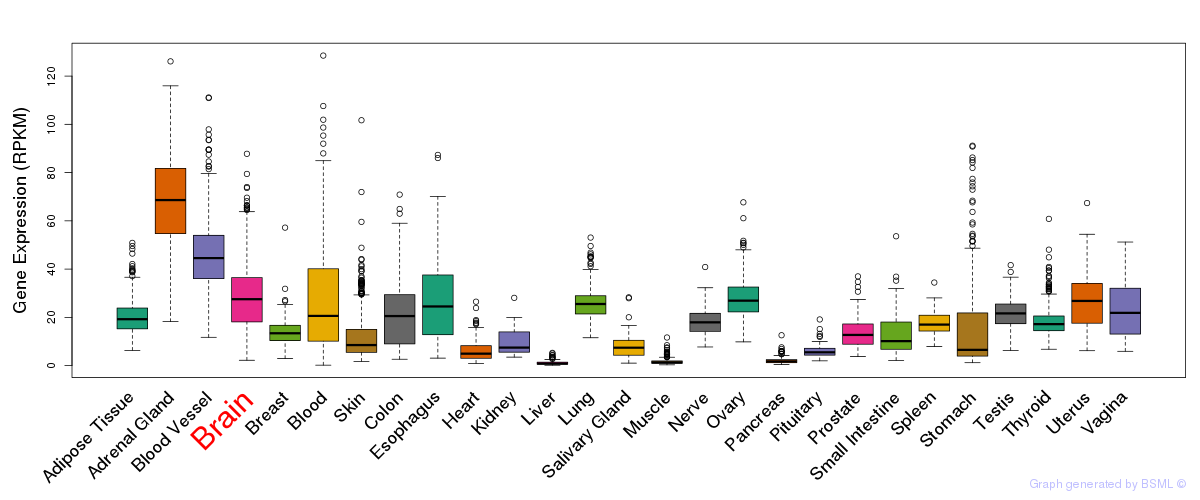
 Differentially methylated gene
Differentially methylated gene eQTL annotation
eQTL annotation