Gene Page: KLF12
Summary ?
| GeneID | 11278 |
| Symbol | KLF12 |
| Synonyms | AP-2rep|AP2REP|HSPC122 |
| Description | Kruppel-like factor 12 |
| Reference | MIM:607531|HGNC:HGNC:6346|Ensembl:ENSG00000118922|HPRD:06336|Vega:OTTHUMG00000017078 |
| Gene type | protein-coding |
| Map location | 13q22 |
| Pascal p-value | 0.002 |
| Sherlock p-value | 0.234 |
| TADA p-value | 0.01 |
| Fetal beta | 1.001 |
| DMG | 1 (# studies) |
| eGene | Myers' cis & trans |
| Support | CompositeSet |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CV:PGCnp | Genome-wide Association Study | GWAS | |
| DMG:Jaffe_2016 | Genome-wide DNA methylation analysis | This dataset includes 2,104 probes/CpGs associated with SZ patients (n=108) compared to 136 controls at Bonferroni-adjusted P < 0.05. | 1 |
| DNM:Xu_2012 | Whole Exome Sequencing analysis | De novo mutations of 4 genes were identified by exome sequencing of 795 samples in this study |
Section I. Genetics and epigenetics annotation
 DNM table
DNM table
| Gene | Chromosome | Position | Ref | Alt | Transcript | AA change | Mutation type | Sift | CG46 | Trait | Study |
|---|---|---|---|---|---|---|---|---|---|---|---|
| KLF12 | chr13 | 74289537 | G | C | NM_007249 | p.332S>C | missense | Schizophrenia | DNM:Xu_2012 |
 Differentially methylated gene
Differentially methylated gene
| Probe | Chromosome | Position | Nearest gene | P (dis) | Beta (dis) | FDR (dis) | Study |
|---|---|---|---|---|---|---|---|
| cg10959786 | 13 | 74708124 | KLF12 | 2.7E-8 | -0.01 | 8.55E-6 | DMG:Jaffe_2016 |
 eQTL annotation
eQTL annotation
| SNP ID | Chromosome | Position | eGene | Gene Entrez ID | pvalue | qvalue | TSS distance | eQTL type |
|---|---|---|---|---|---|---|---|---|
| rs1561519 | chr2 | 35823020 | KLF12 | 11278 | 0.01 | trans | ||
| rs1371418 | chr2 | 35830333 | KLF12 | 11278 | 0.03 | trans | ||
| rs7570146 | chr2 | 35844751 | KLF12 | 11278 | 0.01 | trans | ||
| rs1439689 | chr2 | 35851217 | KLF12 | 11278 | 0.14 | trans | ||
| rs1439691 | chr2 | 35851281 | KLF12 | 11278 | 0.01 | trans | ||
| rs9355872 | chr6 | 161534332 | KLF12 | 11278 | 0.17 | trans | ||
| rs9548306 | chr13 | 38876011 | KLF12 | 11278 | 0.17 | trans |
Section II. Transcriptome annotation
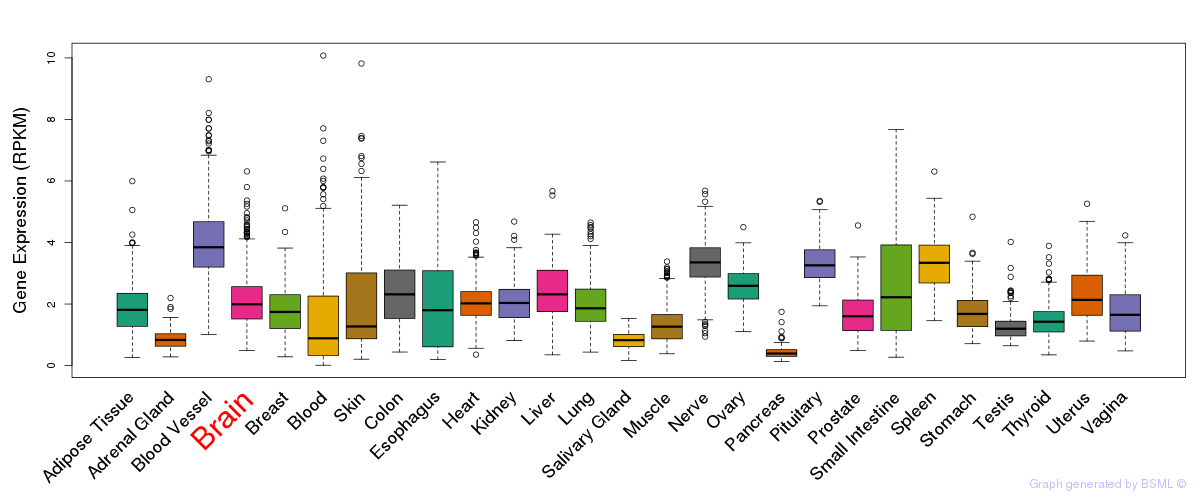
General gene expression (GTEx)
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
No co-expressed genes in brain regions
Section III. Gene Ontology annotation
| Molecular function | GO term | Evidence | Neuro keywords | PubMed ID |
|---|---|---|---|---|
| GO:0003700 | transcription factor activity | IDA | 16615998 | |
| GO:0003714 | transcription corepressor activity | TAS | 10704285 | |
| GO:0008270 | zinc ion binding | IEA | - | |
| GO:0046872 | metal ion binding | IEA | - | |
| Biological process | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0000122 | negative regulation of transcription from RNA polymerase II promoter | IDA | 16615998 | |
| GO:0006350 | transcription | IEA | - | |
| GO:0045944 | positive regulation of transcription from RNA polymerase II promoter | IEA | - | |
| Cellular component | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0005622 | intracellular | IEA | - | |
| GO:0005634 | nucleus | IEA | - |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| PARENT MTOR SIGNALING UP | 567 | 375 | All SZGR 2.0 genes in this pathway |
| ZHONG RESPONSE TO AZACITIDINE AND TSA UP | 183 | 119 | All SZGR 2.0 genes in this pathway |
| ZHOU INFLAMMATORY RESPONSE LIVE DN | 384 | 220 | All SZGR 2.0 genes in this pathway |
| THUM SYSTOLIC HEART FAILURE DN | 244 | 147 | All SZGR 2.0 genes in this pathway |
| ZHOU INFLAMMATORY RESPONSE FIMA UP | 544 | 308 | All SZGR 2.0 genes in this pathway |
| ODONNELL METASTASIS UP | 82 | 58 | All SZGR 2.0 genes in this pathway |
| GAUSSMANN MLL AF4 FUSION TARGETS C UP | 170 | 114 | All SZGR 2.0 genes in this pathway |
| KAN RESPONSE TO ARSENIC TRIOXIDE | 123 | 80 | All SZGR 2.0 genes in this pathway |
| SCHLOSSER SERUM RESPONSE UP | 134 | 93 | All SZGR 2.0 genes in this pathway |
| AMUNDSON RESPONSE TO ARSENITE | 217 | 143 | All SZGR 2.0 genes in this pathway |
| PUJANA ATM PCC NETWORK | 1442 | 892 | All SZGR 2.0 genes in this pathway |
| BUYTAERT PHOTODYNAMIC THERAPY STRESS UP | 811 | 508 | All SZGR 2.0 genes in this pathway |
| SCHAEFFER PROSTATE DEVELOPMENT 48HR DN | 428 | 306 | All SZGR 2.0 genes in this pathway |
| IVANOVA HEMATOPOIESIS STEM CELL AND PROGENITOR | 681 | 420 | All SZGR 2.0 genes in this pathway |
| MASSARWEH TAMOXIFEN RESISTANCE UP | 578 | 341 | All SZGR 2.0 genes in this pathway |
| MELLMAN TUT1 TARGETS DN | 47 | 29 | All SZGR 2.0 genes in this pathway |
| BRUINS UVC RESPONSE VIA TP53 GROUP A | 898 | 516 | All SZGR 2.0 genes in this pathway |
| PURBEY TARGETS OF CTBP1 NOT SATB1 DN | 448 | 282 | All SZGR 2.0 genes in this pathway |
| PECE MAMMARY STEM CELL DN | 146 | 88 | All SZGR 2.0 genes in this pathway |
Section VI. microRNA annotation
| miRNA family | Target position | miRNA ID | miRNA seq | ||
|---|---|---|---|---|---|
| UTR start | UTR end | Match method | |||
| miR-101 | 9108 | 9114 | 1A | hsa-miR-101 | UACAGUACUGUGAUAACUGAAG |
| miR-124.1 | 4018 | 4024 | 1A | hsa-miR-124a | UUAAGGCACGCGGUGAAUGCCA |
| miR-137 | 1565 | 1572 | 1A,m8 | hsa-miR-137 | UAUUGCUUAAGAAUACGCGUAG |
| hsa-miR-137 | UAUUGCUUAAGAAUACGCGUAG | ||||
| hsa-miR-137 | UAUUGCUUAAGAAUACGCGUAG | ||||
| miR-138 | 8339 | 8345 | 1A | hsa-miR-138brain | AGCUGGUGUUGUGAAUC |
| hsa-miR-138brain | AGCUGGUGUUGUGAAUC | ||||
| hsa-miR-138brain | AGCUGGUGUUGUGAAUC | ||||
| miR-141/200a | 9220 | 9226 | 1A | hsa-miR-141 | UAACACUGUCUGGUAAAGAUGG |
| hsa-miR-200a | UAACACUGUCUGGUAACGAUGU | ||||
| miR-144 | 9108 | 9114 | 1A | hsa-miR-144 | UACAGUAUAGAUGAUGUACUAG |
| miR-151 | 8463 | 8470 | 1A,m8 | hsa-miR-151brain | ACUAGACUGAAGCUCCUUGAGG |
| miR-17-5p/20/93.mr/106/519.d | 3468 | 3474 | m8 | hsa-miR-17-5p | CAAAGUGCUUACAGUGCAGGUAGU |
| hsa-miR-20abrain | UAAAGUGCUUAUAGUGCAGGUAG | ||||
| hsa-miR-106a | AAAAGUGCUUACAGUGCAGGUAGC | ||||
| hsa-miR-106bSZ | UAAAGUGCUGACAGUGCAGAU | ||||
| hsa-miR-20bSZ | CAAAGUGCUCAUAGUGCAGGUAG | ||||
| hsa-miR-519d | CAAAGUGCCUCCCUUUAGAGUGU | ||||
| hsa-miR-17-5p | CAAAGUGCUUACAGUGCAGGUAGU | ||||
| hsa-miR-20abrain | UAAAGUGCUUAUAGUGCAGGUAG | ||||
| hsa-miR-106a | AAAAGUGCUUACAGUGCAGGUAGC | ||||
| hsa-miR-106bSZ | UAAAGUGCUGACAGUGCAGAU | ||||
| hsa-miR-20bSZ | CAAAGUGCUCAUAGUGCAGGUAG | ||||
| hsa-miR-519d | CAAAGUGCCUCCCUUUAGAGUGU | ||||
| miR-188 | 1077 | 1083 | m8 | hsa-miR-188 | CAUCCCUUGCAUGGUGGAGGGU |
| miR-194 | 469 | 475 | m8 | hsa-miR-194 | UGUAACAGCAACUCCAUGUGGA |
| miR-199 | 901 | 907 | m8 | hsa-miR-199a | CCCAGUGUUCAGACUACCUGUUC |
| hsa-miR-199b | CCCAGUGUUUAGACUAUCUGUUC | ||||
| miR-200bc/429 | 3740 | 3746 | m8 | hsa-miR-200b | UAAUACUGCCUGGUAAUGAUGAC |
| hsa-miR-200c | UAAUACUGCCGGGUAAUGAUGG | ||||
| hsa-miR-429 | UAAUACUGUCUGGUAAAACCGU | ||||
| hsa-miR-200b | UAAUACUGCCUGGUAAUGAUGAC | ||||
| hsa-miR-200c | UAAUACUGCCGGGUAAUGAUGG | ||||
| hsa-miR-429 | UAAUACUGUCUGGUAAAACCGU | ||||
| miR-203.1 | 2886 | 2892 | 1A | hsa-miR-203 | UGAAAUGUUUAGGACCACUAG |
| miR-204/211 | 1076 | 1082 | m8 | hsa-miR-204brain | UUCCCUUUGUCAUCCUAUGCCU |
| hsa-miR-211 | UUCCCUUUGUCAUCCUUCGCCU | ||||
| miR-205 | 9358 | 9364 | m8 | hsa-miR-205 | UCCUUCAUUCCACCGGAGUCUG |
| hsa-miR-205 | UCCUUCAUUCCACCGGAGUCUG | ||||
| miR-217 | 3737 | 3744 | 1A,m8 | hsa-miR-217 | UACUGCAUCAGGAACUGAUUGGAU |
| miR-218 | 2922 | 2928 | m8 | hsa-miR-218brain | UUGUGCUUGAUCUAACCAUGU |
| hsa-miR-218brain | UUGUGCUUGAUCUAACCAUGU | ||||
| miR-224 | 8441 | 8447 | 1A | hsa-miR-224 | CAAGUCACUAGUGGUUCCGUUUA |
| miR-25/32/92/363/367 | 1745 | 1751 | 1A | hsa-miR-25brain | CAUUGCACUUGUCUCGGUCUGA |
| hsa-miR-32 | UAUUGCACAUUACUAAGUUGC | ||||
| hsa-miR-92 | UAUUGCACUUGUCCCGGCCUG | ||||
| hsa-miR-367 | AAUUGCACUUUAGCAAUGGUGA | ||||
| hsa-miR-92bSZ | UAUUGCACUCGUCCCGGCCUC | ||||
| hsa-miR-25brain | CAUUGCACUUGUCUCGGUCUGA | ||||
| hsa-miR-32 | UAUUGCACAUUACUAAGUUGC | ||||
| hsa-miR-92 | UAUUGCACUUGUCCCGGCCUG | ||||
| hsa-miR-367 | AAUUGCACUUUAGCAAUGGUGA | ||||
| hsa-miR-92bSZ | UAUUGCACUCGUCCCGGCCUC | ||||
| miR-27 | 9301 | 9307 | 1A | hsa-miR-27abrain | UUCACAGUGGCUAAGUUCCGC |
| hsa-miR-27bbrain | UUCACAGUGGCUAAGUUCUGC | ||||
| miR-29 | 1339 | 1346 | 1A,m8 | hsa-miR-29aSZ | UAGCACCAUCUGAAAUCGGUU |
| hsa-miR-29bSZ | UAGCACCAUUUGAAAUCAGUGUU | ||||
| hsa-miR-29cSZ | UAGCACCAUUUGAAAUCGGU | ||||
| miR-329 | 8481 | 8487 | m8 | hsa-miR-329brain | AACACACCUGGUUAACCUCUUU |
| miR-33 | 8294 | 8300 | 1A | hsa-miR-33 | GUGCAUUGUAGUUGCAUUG |
| hsa-miR-33b | GUGCAUUGCUGUUGCAUUGCA | ||||
| miR-34/449 | 4249 | 4255 | m8 | hsa-miR-34abrain | UGGCAGUGUCUUAGCUGGUUGUU |
| hsa-miR-34c | AGGCAGUGUAGUUAGCUGAUUGC | ||||
| hsa-miR-449 | UGGCAGUGUAUUGUUAGCUGGU | ||||
| hsa-miR-449b | AGGCAGUGUAUUGUUAGCUGGC | ||||
| miR-342 | 8227 | 8233 | 1A | hsa-miR-342brain | UCUCACACAGAAAUCGCACCCGUC |
| miR-34b | 4250 | 4256 | m8 | hsa-miR-34b | UAGGCAGUGUCAUUAGCUGAUUG |
| miR-370 | 57 | 63 | m8 | hsa-miR-370brain | GCCUGCUGGGGUGGAACCUGG |
| hsa-miR-370brain | GCCUGCUGGGGUGGAACCUGG | ||||
| miR-376 | 9289 | 9295 | 1A | hsa-miR-376a | AUCAUAGAGGAAAAUCCACGU |
| hsa-miR-376b | AUCAUAGAGGAAAAUCCAUGUU | ||||
| miR-376c | 8180 | 8186 | m8 | hsa-miR-376c | AACAUAGAGGAAAUUCCACG |
| miR-381 | 9053 | 9059 | m8 | hsa-miR-381 | UAUACAAGGGCAAGCUCUCUGU |
| hsa-miR-381 | UAUACAAGGGCAAGCUCUCUGU | ||||
| miR-409-5p | 163 | 169 | m8 | hsa-miR-409-5p | AGGUUACCCGAGCAACUUUGCA |
| miR-410 | 8989 | 8995 | 1A | hsa-miR-410 | AAUAUAACACAGAUGGCCUGU |
| miR-539 | 7353 | 7360 | 1A,m8 | hsa-miR-539 | GGAGAAAUUAUCCUUGGUGUGU |
| miR-542-3p | 825 | 831 | m8 | hsa-miR-542-3p | UGUGACAGAUUGAUAACUGAAA |
| miR-7 | 4218 | 4225 | 1A,m8 | hsa-miR-7SZ | UGGAAGACUAGUGAUUUUGUUG |
| miR-9 | 3776 | 3782 | m8 | hsa-miR-9SZ | UCUUUGGUUAUCUAGCUGUAUGA |
| miR-93.hd/291-3p/294/295/302/372/373/520 | 3467 | 3473 | m8 | hsa-miR-93brain | AAAGUGCUGUUCGUGCAGGUAG |
| hsa-miR-302a | UAAGUGCUUCCAUGUUUUGGUGA | ||||
| hsa-miR-302b | UAAGUGCUUCCAUGUUUUAGUAG | ||||
| hsa-miR-302c | UAAGUGCUUCCAUGUUUCAGUGG | ||||
| hsa-miR-302d | UAAGUGCUUCCAUGUUUGAGUGU | ||||
| hsa-miR-372 | AAAGUGCUGCGACAUUUGAGCGU | ||||
| hsa-miR-373 | GAAGUGCUUCGAUUUUGGGGUGU | ||||
| hsa-miR-520e | AAAGUGCUUCCUUUUUGAGGG | ||||
| hsa-miR-520a | AAAGUGCUUCCCUUUGGACUGU | ||||
| hsa-miR-520b | AAAGUGCUUCCUUUUAGAGGG | ||||
| hsa-miR-520c | AAAGUGCUUCCUUUUAGAGGGUU | ||||
| hsa-miR-520d | AAAGUGCUUCUCUUUGGUGGGUU | ||||
- SZ: miRNAs which differentially expressed in brain cortex of schizophrenia patients comparing with control samples using microarray. Click here to see the list of SZ related miRNAs.
- Brain: miRNAs which are expressed in brain based on miRNA microarray expression studies. Click here to see the list of brain related miRNAs.