Gene Page: CNTFR
Summary ?
| GeneID | 1271 |
| Symbol | CNTFR |
| Synonyms | - |
| Description | ciliary neurotrophic factor receptor |
| Reference | MIM:118946|HGNC:HGNC:2170|Ensembl:ENSG00000122756|HPRD:00348|Vega:OTTHUMG00000019821 |
| Gene type | protein-coding |
| Map location | 9p13 |
| Pascal p-value | 0.045 |
| Sherlock p-value | 5.53E-5 |
| Fetal beta | -0.293 |
| eGene | Caudate basal ganglia Myers' cis & trans |
| Support | G2Cdb.humanPSD G2Cdb.humanPSP |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CV:PGCnp | Genome-wide Association Study | GWAS | |
| GO_Annotation | Mapping neuro-related keywords to Gene Ontology annotations | Hits with neuro-related keywords: 2 |
Section I. Genetics and epigenetics annotation
 eQTL annotation
eQTL annotation
| SNP ID | Chromosome | Position | eGene | Gene Entrez ID | pvalue | qvalue | TSS distance | eQTL type |
|---|---|---|---|---|---|---|---|---|
| rs11006200 | chr10 | 52339779 | CNTFR | 1271 | 0.17 | trans |
Section II. Transcriptome annotation
General gene expression (GTEx)
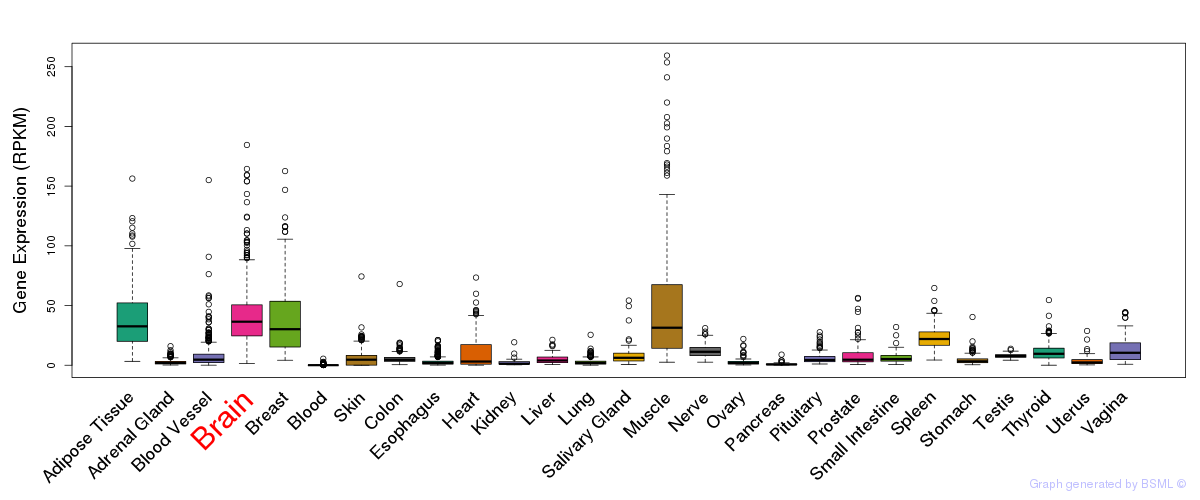
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
Top co-expressed genes in brain regions
| Top 10 positively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| TNFRSF19 | 0.74 | 0.65 |
| DCT | 0.68 | -0.02 |
| MSTN | 0.67 | 0.29 |
| PDLIM1 | 0.67 | 0.40 |
| NEUROD1 | 0.66 | 0.49 |
| FGFR1 | 0.66 | 0.34 |
| BOC | 0.65 | 0.42 |
| JUB | 0.64 | 0.43 |
| COL4A6 | 0.63 | 0.60 |
| LTBP1 | 0.63 | 0.29 |
| Top 10 negatively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| C5orf53 | -0.19 | -0.55 |
| MGLL | -0.18 | -0.30 |
| ASPHD1 | -0.18 | -0.45 |
| CCNI2 | -0.18 | -0.46 |
| FBXO2 | -0.18 | -0.41 |
| HLA-F | -0.18 | -0.48 |
| SLC16A11 | -0.18 | -0.41 |
| INPP1 | -0.18 | -0.07 |
| TMEM54 | -0.18 | -0.39 |
| APOL1 | -0.17 | -0.49 |
Section III. Gene Ontology annotation
| Molecular function | GO term | Evidence | Neuro keywords | PubMed ID |
|---|---|---|---|---|
| GO:0004897 | ciliary neurotrophic factor receptor activity | TAS | neurotrophic (GO term level: 7) | 7585948 |
| GO:0004872 | receptor activity | IEA | - | |
| GO:0004896 | cytokine receptor activity | IEA | - | |
| GO:0005515 | protein binding | IEA | - | |
| GO:0019955 | cytokine binding | IPI | 15272019 | |
| Biological process | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0007399 | nervous system development | TAS | neurite (GO term level: 5) | 7585948 |
| GO:0007165 | signal transduction | NAS | 1648265 | |
| GO:0008284 | positive regulation of cell proliferation | IEA | - | |
| Cellular component | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0016020 | membrane | IEA | - | |
| GO:0005886 | plasma membrane | IEA | - | |
| GO:0031225 | anchored to membrane | IEA | - | |
| GO:0019898 | extrinsic to membrane | TAS | 1648265 |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| KEGG CYTOKINE CYTOKINE RECEPTOR INTERACTION | 267 | 161 | All SZGR 2.0 genes in this pathway |
| KEGG JAK STAT SIGNALING PATHWAY | 155 | 105 | All SZGR 2.0 genes in this pathway |
| GRAESSMANN APOPTOSIS BY SERUM DEPRIVATION UP | 552 | 347 | All SZGR 2.0 genes in this pathway |
| BENPORATH SUZ12 TARGETS | 1038 | 678 | All SZGR 2.0 genes in this pathway |
| BENPORATH EED TARGETS | 1062 | 725 | All SZGR 2.0 genes in this pathway |
| BENPORATH ES WITH H3K27ME3 | 1118 | 744 | All SZGR 2.0 genes in this pathway |
| BENPORATH PRC2 TARGETS | 652 | 441 | All SZGR 2.0 genes in this pathway |
| BRUINS UVC RESPONSE VIA TP53 GROUP A | 898 | 516 | All SZGR 2.0 genes in this pathway |
| PEDRIOLI MIR31 TARGETS UP | 221 | 120 | All SZGR 2.0 genes in this pathway |
Section VI. microRNA annotation
| miRNA family | Target position | miRNA ID | miRNA seq | ||
|---|---|---|---|---|---|
| UTR start | UTR end | Match method | |||
| miR-146 | 611 | 617 | 1A | hsa-miR-146a | UGAGAACUGAAUUCCAUGGGUU |
| hsa-miR-146bbrain | UGAGAACUGAAUUCCAUAGGCU | ||||
| miR-19 | 82 | 89 | 1A,m8 | hsa-miR-19a | UGUGCAAAUCUAUGCAAAACUGA |
| hsa-miR-19b | UGUGCAAAUCCAUGCAAAACUGA | ||||
| miR-200bc/429 | 510 | 516 | m8 | hsa-miR-200b | UAAUACUGCCUGGUAAUGAUGAC |
| hsa-miR-200c | UAAUACUGCCGGGUAAUGAUGG | ||||
| hsa-miR-429 | UAAUACUGUCUGGUAAAACCGU | ||||
| miR-203.1 | 629 | 635 | 1A | hsa-miR-203 | UGAAAUGUUUAGGACCACUAG |
| miR-21 | 392 | 398 | m8 | hsa-miR-21brain | UAGCUUAUCAGACUGAUGUUGA |
| hsa-miR-590 | GAGCUUAUUCAUAAAAGUGCAG | ||||
| miR-22 | 219 | 225 | 1A | hsa-miR-22brain | AAGCUGCCAGUUGAAGAACUGU |
| miR-24 | 56 | 62 | m8 | hsa-miR-24SZ | UGGCUCAGUUCAGCAGGAACAG |
| miR-28 | 395 | 401 | m8 | hsa-miR-28brain | AAGGAGCUCACAGUCUAUUGAG |
| miR-370 | 41 | 47 | 1A | hsa-miR-370brain | GCCUGCUGGGGUGGAACCUGG |
| miR-374 | 389 | 395 | 1A | hsa-miR-374 | UUAUAAUACAACCUGAUAAGUG |
| miR-485-5p | 348 | 354 | m8 | hsa-miR-485-5p | AGAGGCUGGCCGUGAUGAAUUC |
| miR-504 | 365 | 371 | m8 | hsa-miR-504 | AGACCCUGGUCUGCACUCUAU |
| miR-9 | 234 | 241 | 1A,m8 | hsa-miR-9SZ | UCUUUGGUUAUCUAGCUGUAUGA |
- SZ: miRNAs which differentially expressed in brain cortex of schizophrenia patients comparing with control samples using microarray. Click here to see the list of SZ related miRNAs.
- Brain: miRNAs which are expressed in brain based on miRNA microarray expression studies. Click here to see the list of brain related miRNAs.