Gene Page: CR1
Summary ?
| GeneID | 1378 |
| Symbol | CR1 |
| Synonyms | C3BR|C4BR|CD35|KN |
| Description | complement component 3b/4b receptor 1 (Knops blood group) |
| Reference | MIM:120620|HGNC:HGNC:2334|Ensembl:ENSG00000203710|HPRD:00398|Vega:OTTHUMG00000036311 |
| Gene type | protein-coding |
| Map location | 1q32 |
| Fetal beta | -0.091 |
| DMG | 1 (# studies) |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CV:PGCnp | Genome-wide Association Study | GWAS | |
| DMG:vanEijk_2014 | Genome-wide DNA methylation analysis | This dataset includes 432 differentially methylated CpG sites corresponding to 391 unique transcripts between schizophrenia patients (n=260) and unaffected controls (n=250). | 4 |
| PMID:cooccur | High-throughput literature-search | Systematic search in PubMed for genes co-occurring with SCZ keywords. A total of 3027 genes were included. | |
| Literature | High-throughput literature-search | Co-occurance with Schizophrenia keywords: schizophrenia,schizophrenias | Click to show details |
Section I. Genetics and epigenetics annotation
 Differentially methylated gene
Differentially methylated gene
| Probe | Chromosome | Position | Nearest gene | P (dis) | Beta (dis) | FDR (dis) | Study |
|---|---|---|---|---|---|---|---|
| cg14726637 | 1 | 207669308 | CR1 | 5.098E-4 | -3.744 | DMG:vanEijk_2014 | |
| cg14726637 | 1 | 207669308 | CR1 | 1.326E-4 | -4.929 | DMG:vanEijk_2014 | |
| cg03583857 | 1 | 208085022 | CR1 | 1.067E-4 | -5.665 | DMG:vanEijk_2014 | |
| cg03583857 | 1 | 208085022 | CR1 | 8.46E-5 | -6.904 | DMG:vanEijk_2014 |
Section II. Transcriptome annotation
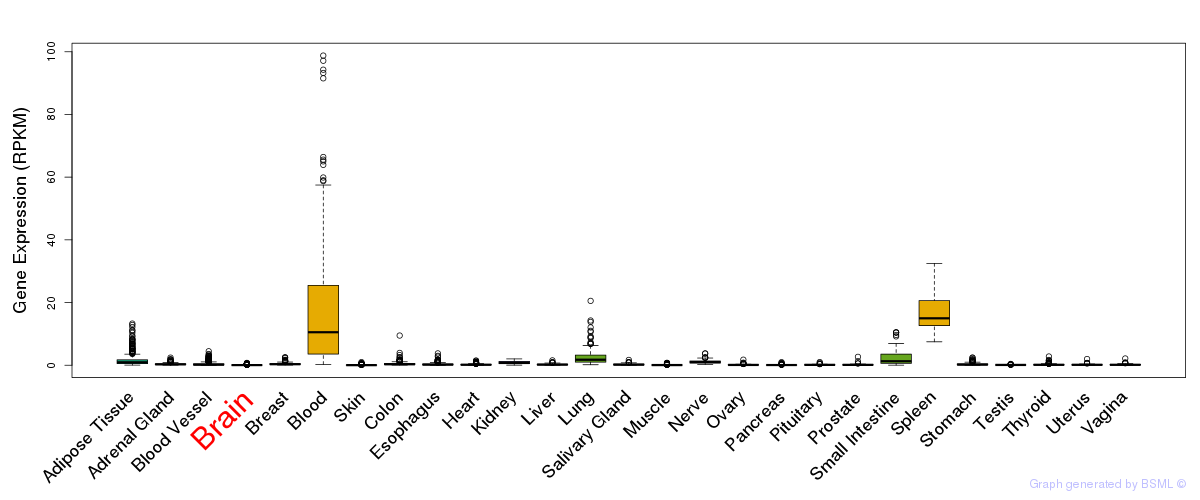
General gene expression (GTEx)
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
Top co-expressed genes in brain regions
| Top 10 positively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| ARID1A | 0.98 | 0.99 |
| EP300 | 0.98 | 0.98 |
| ZMIZ1 | 0.98 | 0.98 |
| TRRAP | 0.98 | 0.98 |
| SETD1B | 0.98 | 0.99 |
| PHF12 | 0.97 | 0.97 |
| EP400 | 0.97 | 0.97 |
| HCFC1 | 0.97 | 0.97 |
| POGZ | 0.97 | 0.96 |
| C11orf30 | 0.97 | 0.95 |
| Top 10 negatively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| AF347015.31 | -0.70 | -0.87 |
| C5orf53 | -0.70 | -0.77 |
| S100B | -0.70 | -0.83 |
| FXYD1 | -0.69 | -0.84 |
| MT-CO2 | -0.69 | -0.86 |
| AF347015.27 | -0.67 | -0.82 |
| SERPINB6 | -0.67 | -0.73 |
| IFI27 | -0.67 | -0.81 |
| AIFM3 | -0.67 | -0.71 |
| AF347015.33 | -0.67 | -0.80 |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| KEGG COMPLEMENT AND COAGULATION CASCADES | 69 | 49 | All SZGR 2.0 genes in this pathway |
| KEGG HEMATOPOIETIC CELL LINEAGE | 88 | 60 | All SZGR 2.0 genes in this pathway |
| KEGG LEISHMANIA INFECTION | 72 | 56 | All SZGR 2.0 genes in this pathway |
| BIOCARTA BLYMPHOCYTE PATHWAY | 11 | 8 | All SZGR 2.0 genes in this pathway |
| REACTOME REGULATION OF COMPLEMENT CASCADE | 14 | 12 | All SZGR 2.0 genes in this pathway |
| REACTOME INNATE IMMUNE SYSTEM | 279 | 178 | All SZGR 2.0 genes in this pathway |
| REACTOME IMMUNE SYSTEM | 933 | 616 | All SZGR 2.0 genes in this pathway |
| REACTOME COMPLEMENT CASCADE | 32 | 22 | All SZGR 2.0 genes in this pathway |
| SENGUPTA NASOPHARYNGEAL CARCINOMA DN | 349 | 157 | All SZGR 2.0 genes in this pathway |
| DEURIG T CELL PROLYMPHOCYTIC LEUKEMIA UP | 368 | 234 | All SZGR 2.0 genes in this pathway |
| OSWALD HEMATOPOIETIC STEM CELL IN COLLAGEN GEL DN | 275 | 168 | All SZGR 2.0 genes in this pathway |
| TAKEDA TARGETS OF NUP98 HOXA9 FUSION 8D DN | 205 | 127 | All SZGR 2.0 genes in this pathway |
| KIM WT1 TARGETS 8HR DN | 129 | 84 | All SZGR 2.0 genes in this pathway |
| DODD NASOPHARYNGEAL CARCINOMA UP | 1821 | 933 | All SZGR 2.0 genes in this pathway |
| PUJANA ATM PCC NETWORK | 1442 | 892 | All SZGR 2.0 genes in this pathway |
| TARTE PLASMA CELL VS PLASMABLAST UP | 398 | 262 | All SZGR 2.0 genes in this pathway |
| LENAOUR DENDRITIC CELL MATURATION UP | 114 | 84 | All SZGR 2.0 genes in this pathway |
| MOREAUX MULTIPLE MYELOMA BY TACI UP | 412 | 249 | All SZGR 2.0 genes in this pathway |
| BLALOCK ALZHEIMERS DISEASE INCIPIENT UP | 390 | 242 | All SZGR 2.0 genes in this pathway |
| BLALOCK ALZHEIMERS DISEASE UP | 1691 | 1088 | All SZGR 2.0 genes in this pathway |
| BAELDE DIABETIC NEPHROPATHY DN | 434 | 302 | All SZGR 2.0 genes in this pathway |
| KRIGE RESPONSE TO TOSEDOSTAT 6HR UP | 953 | 554 | All SZGR 2.0 genes in this pathway |
| KRIGE RESPONSE TO TOSEDOSTAT 24HR UP | 783 | 442 | All SZGR 2.0 genes in this pathway |
| BONOME OVARIAN CANCER SURVIVAL SUBOPTIMAL DEBULKING | 510 | 309 | All SZGR 2.0 genes in this pathway |
| STAMBOLSKY RESPONSE TO VITAMIN D3 UP | 84 | 48 | All SZGR 2.0 genes in this pathway |
| WIERENGA STAT5A TARGETS DN | 213 | 127 | All SZGR 2.0 genes in this pathway |