Gene Page: CRABP1
Summary ?
| GeneID | 1381 |
| Symbol | CRABP1 |
| Synonyms | CRABP|CRABP-I|CRABPI|RBP5 |
| Description | cellular retinoic acid binding protein 1 |
| Reference | MIM:180230|HGNC:HGNC:2338|Ensembl:ENSG00000166426|HPRD:07520|Vega:OTTHUMG00000143862 |
| Gene type | protein-coding |
| Map location | 15q24 |
| Pascal p-value | 0.085 |
| Sherlock p-value | 0.004 |
| Fetal beta | -0.266 |
| eGene | Myers' cis & trans |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CV:PGCnp | Genome-wide Association Study | GWAS |
Section I. Genetics and epigenetics annotation
 eQTL annotation
eQTL annotation
| SNP ID | Chromosome | Position | eGene | Gene Entrez ID | pvalue | qvalue | TSS distance | eQTL type |
|---|---|---|---|---|---|---|---|---|
| rs17131472 | chr1 | 91962966 | CRABP1 | 1381 | 0.12 | trans | ||
| rs17535450 | chr2 | 39900012 | CRABP1 | 1381 | 0.03 | trans | ||
| rs11706314 | chr3 | 45309716 | CRABP1 | 1381 | 0 | trans | ||
| rs11706394 | chr3 | 45309907 | CRABP1 | 1381 | 0.01 | trans | ||
| rs2922180 | chr3 | 125280472 | CRABP1 | 1381 | 0 | trans | ||
| rs2976809 | chr3 | 125451738 | CRABP1 | 1381 | 5.004E-4 | trans | ||
| rs17090446 | chr4 | 62255221 | CRABP1 | 1381 | 0.15 | trans | ||
| rs4241605 | chr4 | 78161505 | CRABP1 | 1381 | 0.19 | trans | ||
| rs7668550 | chr4 | 104384355 | CRABP1 | 1381 | 0.02 | trans | ||
| rs3796892 | chr4 | 111404774 | CRABP1 | 1381 | 0.09 | trans | ||
| rs16894149 | chr5 | 61013724 | CRABP1 | 1381 | 0.02 | trans | ||
| rs6449550 | chr5 | 61034312 | CRABP1 | 1381 | 0.01 | trans | ||
| rs4489051 | chr5 | 147954484 | CRABP1 | 1381 | 0.02 | trans | ||
| rs6874540 | chr5 | 149911669 | CRABP1 | 1381 | 0.15 | trans | ||
| rs2273232 | chr5 | 149912526 | CRABP1 | 1381 | 0.15 | trans | ||
| rs17056435 | chr5 | 158453782 | CRABP1 | 1381 | 0.02 | trans | ||
| rs9313797 | chr5 | 158465457 | CRABP1 | 1381 | 0.01 | trans | ||
| rs17056474 | chr5 | 158468105 | CRABP1 | 1381 | 0.01 | trans | ||
| rs2294262 | chr6 | 16108966 | CRABP1 | 1381 | 0.11 | trans | ||
| rs6455226 | chr6 | 67930295 | CRABP1 | 1381 | 0.19 | trans | ||
| rs16868852 | chr6 | 70980333 | CRABP1 | 1381 | 0.2 | trans | ||
| rs9387571 | chr6 | 118600137 | CRABP1 | 1381 | 0.08 | trans | ||
| rs283063 | chr6 | 118621313 | CRABP1 | 1381 | 0.04 | trans | ||
| rs9321275 | chr6 | 131493217 | CRABP1 | 1381 | 0.08 | trans | ||
| rs2430483 | chr7 | 104548631 | CRABP1 | 1381 | 0.04 | trans | ||
| rs2199402 | chr8 | 9201002 | CRABP1 | 1381 | 0.01 | trans | ||
| rs7841407 | chr8 | 9243427 | CRABP1 | 1381 | 3.327E-4 | trans | ||
| rs2410231 | chr8 | 14954549 | CRABP1 | 1381 | 0.18 | trans | ||
| rs7029518 | chr9 | 37782560 | CRABP1 | 1381 | 0.11 | trans | ||
| rs10521469 | chr9 | 78884328 | CRABP1 | 1381 | 0.13 | trans | ||
| rs998410 | chr9 | 117622673 | CRABP1 | 1381 | 0.07 | trans | ||
| rs9423711 | chr10 | 2574802 | CRABP1 | 1381 | 0.01 | trans | ||
| rs748375 | chr10 | 33440178 | CRABP1 | 1381 | 0.14 | trans | ||
| rs11820030 | chr11 | 76133564 | CRABP1 | 1381 | 0 | trans | ||
| rs17134872 | chr11 | 76139845 | CRABP1 | 1381 | 1.674E-4 | trans | ||
| rs10751255 | 0 | CRABP1 | 1381 | 0.01 | trans | |||
| rs11236774 | chr11 | 76234952 | CRABP1 | 1381 | 1.674E-4 | trans | ||
| rs11067174 | chr12 | 115011927 | CRABP1 | 1381 | 0.08 | trans | ||
| rs4349050 | chr13 | 41958722 | CRABP1 | 1381 | 0.19 | trans | ||
| rs7982172 | chr13 | 41969697 | CRABP1 | 1381 | 0.16 | trans | ||
| rs4942043 | chr13 | 41978218 | CRABP1 | 1381 | 0.13 | trans | ||
| rs1750009 | chr13 | 42657093 | CRABP1 | 1381 | 0.02 | trans | ||
| rs9596788 | chr13 | 53924430 | CRABP1 | 1381 | 0.04 | trans | ||
| rs9529974 | chr13 | 72821935 | CRABP1 | 1381 | 3.842E-4 | trans | ||
| rs17720221 | chr13 | 75182377 | CRABP1 | 1381 | 0.03 | trans | ||
| rs17073665 | chr13 | 81541961 | CRABP1 | 1381 | 0.07 | trans | ||
| rs11840833 | chr13 | 94378306 | CRABP1 | 1381 | 0.14 | trans | ||
| rs10149864 | chr14 | 86719683 | CRABP1 | 1381 | 0.1 | trans | ||
| rs2093759 | chr14 | 97171024 | CRABP1 | 1381 | 0.01 | trans | ||
| rs17244419 | chr14 | 97171074 | CRABP1 | 1381 | 0 | trans | ||
| rs7165622 | chr15 | 93979910 | CRABP1 | 1381 | 0.11 | trans | ||
| rs17294918 | chr16 | 52717122 | CRABP1 | 1381 | 0.15 | trans | ||
| rs11873703 | chr18 | 61448702 | CRABP1 | 1381 | 2.158E-5 | trans | ||
| rs17191809 | chr21 | 36444778 | CRABP1 | 1381 | 0.03 | trans | ||
| rs6000401 | chr22 | 37149335 | CRABP1 | 1381 | 0.16 | trans | ||
| rs5991662 | chrX | 43335796 | CRABP1 | 1381 | 0.19 | trans |
Section II. Transcriptome annotation
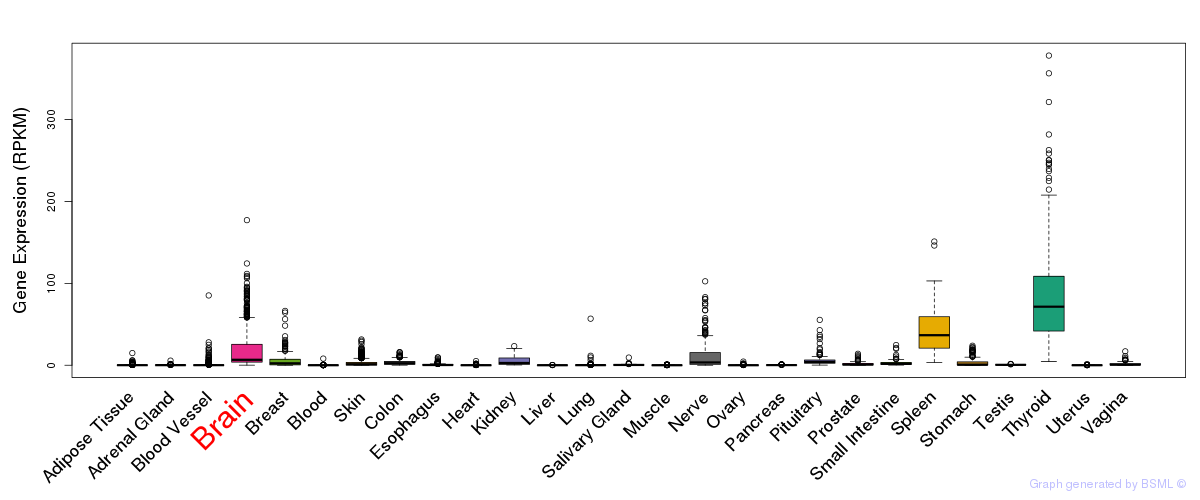
General gene expression (GTEx)
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
Top co-expressed genes in brain regions
| Top 10 positively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| FAM162A | 0.82 | 0.83 |
| ACYP2 | 0.81 | 0.80 |
| TM2D1 | 0.78 | 0.75 |
| NDUFB5 | 0.77 | 0.74 |
| MOCS2 | 0.76 | 0.73 |
| ACOT13 | 0.75 | 0.80 |
| TMEM126B | 0.75 | 0.75 |
| CISD1 | 0.75 | 0.73 |
| NDUFA4 | 0.75 | 0.77 |
| NDUFA9 | 0.75 | 0.72 |
| Top 10 negatively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| VAV2 | -0.57 | -0.56 |
| KIAA1211 | -0.57 | -0.55 |
| VASH1 | -0.57 | -0.57 |
| C21orf30 | -0.57 | -0.60 |
| NHSL1 | -0.57 | -0.64 |
| KIAA1949 | -0.57 | -0.56 |
| SH3BP2 | -0.57 | -0.66 |
| GMIP | -0.57 | -0.47 |
| ANO8 | -0.56 | -0.59 |
| LSS | -0.56 | -0.59 |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| LIU PROSTATE CANCER DN | 481 | 290 | All SZGR 2.0 genes in this pathway |
| LEE NEURAL CREST STEM CELL UP | 146 | 99 | All SZGR 2.0 genes in this pathway |
| KINSEY TARGETS OF EWSR1 FLII FUSION DN | 329 | 219 | All SZGR 2.0 genes in this pathway |
| DELYS THYROID CANCER DN | 232 | 154 | All SZGR 2.0 genes in this pathway |
| CHIARADONNA NEOPLASTIC TRANSFORMATION KRAS CDC25 UP | 58 | 39 | All SZGR 2.0 genes in this pathway |
| CHIARADONNA NEOPLASTIC TRANSFORMATION KRAS UP | 126 | 72 | All SZGR 2.0 genes in this pathway |
| CHIARADONNA NEOPLASTIC TRANSFORMATION CDC25 UP | 120 | 73 | All SZGR 2.0 genes in this pathway |
| MARKEY RB1 CHRONIC LOF UP | 115 | 78 | All SZGR 2.0 genes in this pathway |
| MARKEY RB1 ACUTE LOF DN | 228 | 137 | All SZGR 2.0 genes in this pathway |
| TANAKA METHYLATED IN ESOPHAGEAL CARCINOMA | 103 | 58 | All SZGR 2.0 genes in this pathway |
| LA MEN1 TARGETS | 24 | 15 | All SZGR 2.0 genes in this pathway |
| SCHAEFFER PROSTATE DEVELOPMENT 48HR DN | 428 | 306 | All SZGR 2.0 genes in this pathway |
| RICKMAN HEAD AND NECK CANCER A | 100 | 63 | All SZGR 2.0 genes in this pathway |
| BENPORATH ES 1 | 379 | 235 | All SZGR 2.0 genes in this pathway |
| BENPORATH SUZ12 TARGETS | 1038 | 678 | All SZGR 2.0 genes in this pathway |
| BECKER TAMOXIFEN RESISTANCE UP | 50 | 36 | All SZGR 2.0 genes in this pathway |
| BHATTACHARYA EMBRYONIC STEM CELL | 89 | 60 | All SZGR 2.0 genes in this pathway |
| AFFAR YY1 TARGETS DN | 234 | 137 | All SZGR 2.0 genes in this pathway |
| RODWELL AGING KIDNEY NO BLOOD DN | 150 | 93 | All SZGR 2.0 genes in this pathway |
| RODWELL AGING KIDNEY DN | 145 | 88 | All SZGR 2.0 genes in this pathway |
| YAMAZAKI TCEB3 TARGETS DN | 215 | 132 | All SZGR 2.0 genes in this pathway |
| JACKSON DNMT1 TARGETS DN | 25 | 21 | All SZGR 2.0 genes in this pathway |
| BANDRES RESPONSE TO CARMUSTIN MGMT 48HR DN | 161 | 105 | All SZGR 2.0 genes in this pathway |
| CHEN ETV5 TARGETS TESTIS | 23 | 14 | All SZGR 2.0 genes in this pathway |
| DOUGLAS BMI1 TARGETS UP | 566 | 371 | All SZGR 2.0 genes in this pathway |
| KONDO PROSTATE CANCER HCP WITH H3K27ME3 | 97 | 72 | All SZGR 2.0 genes in this pathway |
| MASSARWEH TAMOXIFEN RESISTANCE DN | 258 | 160 | All SZGR 2.0 genes in this pathway |
| SMID BREAST CANCER LUMINAL B DN | 564 | 326 | All SZGR 2.0 genes in this pathway |
| SMID BREAST CANCER BASAL UP | 648 | 398 | All SZGR 2.0 genes in this pathway |
| MEISSNER NPC HCP WITH H3K4ME2 AND H3K27ME3 | 349 | 234 | All SZGR 2.0 genes in this pathway |
| MEISSNER BRAIN HCP WITH H3K4ME2 AND H3K27ME3 | 59 | 35 | All SZGR 2.0 genes in this pathway |
| HOSHIDA LIVER CANCER SUBCLASS S3 | 266 | 180 | All SZGR 2.0 genes in this pathway |
| NAKAYAMA SOFT TISSUE TUMORS PCA1 DN | 74 | 47 | All SZGR 2.0 genes in this pathway |
| NAKAYAMA SOFT TISSUE TUMORS PCA2 UP | 87 | 50 | All SZGR 2.0 genes in this pathway |
| YAO TEMPORAL RESPONSE TO PROGESTERONE CLUSTER 16 | 79 | 47 | All SZGR 2.0 genes in this pathway |
| MARTENS TRETINOIN RESPONSE UP | 857 | 456 | All SZGR 2.0 genes in this pathway |
| BRUINS UVC RESPONSE LATE | 1137 | 655 | All SZGR 2.0 genes in this pathway |
| VANDESLUIS COMMD1 TARGETS GROUP 3 DN | 39 | 21 | All SZGR 2.0 genes in this pathway |
| FOSTER KDM1A TARGETS UP | 266 | 142 | All SZGR 2.0 genes in this pathway |
| BOSCO TH1 CYTOTOXIC MODULE | 114 | 62 | All SZGR 2.0 genes in this pathway |
| ACEVEDO FGFR1 TARGETS IN PROSTATE CANCER MODEL DN | 308 | 187 | All SZGR 2.0 genes in this pathway |
| DURAND STROMA S UP | 297 | 194 | All SZGR 2.0 genes in this pathway |