Gene Page: ADORA3
Summary ?
| GeneID | 140 |
| Symbol | ADORA3 |
| Synonyms | A3AR |
| Description | adenosine A3 receptor |
| Reference | MIM:600445|HGNC:HGNC:268|Ensembl:ENSG00000121933|Ensembl:ENSG00000282608|HPRD:08984|HPRD:12424|Vega:OTTHUMG00000011957 |
| Gene type | protein-coding |
| Map location | 1p13.2 |
| Pascal p-value | 0.455 |
| Sherlock p-value | 0.898 |
| Fetal beta | -2.573 |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CV:PGCnp | Genome-wide Association Study | GWAS | |
| PMID:cooccur | High-throughput literature-search | Systematic search in PubMed for genes co-occurring with SCZ keywords. A total of 3027 genes were included. | |
| GSMA_I | Genome scan meta-analysis | Psr: 0.0235 | |
| Literature | High-throughput literature-search | Co-occurance with Schizophrenia keywords: schizophrenia,schizophrenias | Click to show details |
Section I. Genetics and epigenetics annotation
Section II. Transcriptome annotation
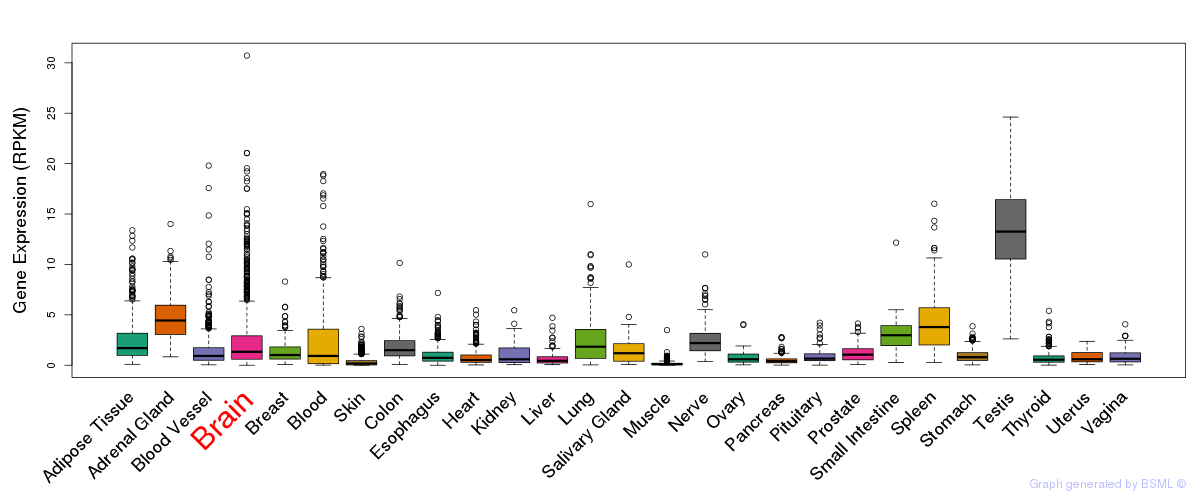
General gene expression (GTEx)
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
Top co-expressed genes in brain regions
| Top 10 positively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| HEATR5A | 0.81 | 0.76 |
| SEPT10 | 0.78 | 0.72 |
| SEPT2 | 0.78 | 0.72 |
| SALL1 | 0.76 | 0.80 |
| CTNNA1 | 0.76 | 0.66 |
| RFTN2 | 0.76 | 0.76 |
| MSN | 0.76 | 0.67 |
| ARHGEF6 | 0.75 | 0.76 |
| NOTCH2 | 0.75 | 0.76 |
| ITGB1 | 0.74 | 0.71 |
| Top 10 negatively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| SLC26A4 | -0.31 | -0.28 |
| DDTL | -0.28 | -0.25 |
| RPL9 | -0.28 | -0.32 |
| A1BG | -0.27 | -0.30 |
| NANOS3 | -0.27 | -0.29 |
| NDUFB1 | -0.27 | -0.22 |
| TTC9B | -0.27 | -0.24 |
| UQCR | -0.26 | -0.28 |
| C17orf37 | -0.26 | -0.28 |
| USMG5 | -0.26 | -0.20 |
Section III. Gene Ontology annotation
| Molecular function | GO term | Evidence | Neuro keywords | PubMed ID |
|---|---|---|---|---|
| GO:0001609 | adenosine receptor activity, G-protein coupled | IEA | - | |
| GO:0004872 | receptor activity | IEA | - | |
| Biological process | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0007190 | activation of adenylate cyclase activity | TAS | 9380026 | |
| GO:0007186 | G-protein coupled receptor protein signaling pathway | IEA | - | |
| GO:0007165 | signal transduction | TAS | 8399349 |9380026 | |
| GO:0008016 | regulation of heart contraction | TAS | 9837869 | |
| GO:0006954 | inflammatory response | TAS | 9164961 | |
| Cellular component | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0005886 | plasma membrane | IEA | - | |
| GO:0005887 | integral to plasma membrane | TAS | 8399349 |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| KEGG NEUROACTIVE LIGAND RECEPTOR INTERACTION | 272 | 195 | All SZGR 2.0 genes in this pathway |
| REACTOME SIGNALING BY GPCR | 920 | 449 | All SZGR 2.0 genes in this pathway |
| REACTOME CLASS A1 RHODOPSIN LIKE RECEPTORS | 305 | 177 | All SZGR 2.0 genes in this pathway |
| REACTOME NUCLEOTIDE LIKE PURINERGIC RECEPTORS | 16 | 8 | All SZGR 2.0 genes in this pathway |
| REACTOME GPCR DOWNSTREAM SIGNALING | 805 | 368 | All SZGR 2.0 genes in this pathway |
| REACTOME G ALPHA I SIGNALLING EVENTS | 195 | 114 | All SZGR 2.0 genes in this pathway |
| REACTOME GPCR LIGAND BINDING | 408 | 246 | All SZGR 2.0 genes in this pathway |
| SCHUETZ BREAST CANCER DUCTAL INVASIVE UP | 351 | 230 | All SZGR 2.0 genes in this pathway |
| TAKEDA TARGETS OF NUP98 HOXA9 FUSION 8D DN | 205 | 127 | All SZGR 2.0 genes in this pathway |
| TAKEDA TARGETS OF NUP98 HOXA9 FUSION 16D DN | 143 | 83 | All SZGR 2.0 genes in this pathway |
| DODD NASOPHARYNGEAL CARCINOMA DN | 1375 | 806 | All SZGR 2.0 genes in this pathway |
| DELYS THYROID CANCER UP | 443 | 294 | All SZGR 2.0 genes in this pathway |
| TSAI RESPONSE TO IONIZING RADIATION | 149 | 101 | All SZGR 2.0 genes in this pathway |
| TARTE PLASMA CELL VS PLASMABLAST UP | 398 | 262 | All SZGR 2.0 genes in this pathway |
| NAKAJIMA EOSINOPHIL | 30 | 20 | All SZGR 2.0 genes in this pathway |
| NIELSEN SCHWANNOMA UP | 18 | 16 | All SZGR 2.0 genes in this pathway |
| KRIGE RESPONSE TO TOSEDOSTAT 6HR DN | 911 | 527 | All SZGR 2.0 genes in this pathway |
| KRIGE RESPONSE TO TOSEDOSTAT 24HR UP | 783 | 442 | All SZGR 2.0 genes in this pathway |
| YOSHIMURA MAPK8 TARGETS UP | 1305 | 895 | All SZGR 2.0 genes in this pathway |
| SHEDDEN LUNG CANCER GOOD SURVIVAL A12 | 317 | 177 | All SZGR 2.0 genes in this pathway |
| MARTENS BOUND BY PML RARA FUSION | 456 | 287 | All SZGR 2.0 genes in this pathway |
| BOSCO TH1 CYTOTOXIC MODULE | 114 | 62 | All SZGR 2.0 genes in this pathway |