Gene Page: PARP4
Summary ?
| GeneID | 143 |
| Symbol | PARP4 |
| Synonyms | ADPRTL1|ARTD4|PARP-4|PARPL|PH5P|VAULT3|VPARP|VWA5C|p193 |
| Description | poly(ADP-ribose) polymerase family member 4 |
| Reference | MIM:607519|HGNC:HGNC:271|Ensembl:ENSG00000102699|HPRD:09598|Vega:OTTHUMG00000016582 |
| Gene type | protein-coding |
| Map location | 13q11 |
| Pascal p-value | 0.189 |
| Fetal beta | 0.343 |
| DMG | 1 (# studies) |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CV:GWASdb | Genome-wide Association Studies | GWASdb records for schizophrenia | |
| CV:PGCnp | Genome-wide Association Study | GWAS | |
| DMG:Jaffe_2016 | Genome-wide DNA methylation analysis | This dataset includes 2,104 probes/CpGs associated with SZ patients (n=108) compared to 136 controls at Bonferroni-adjusted P < 0.05. | 1 |
Section I. Genetics and epigenetics annotation
 Differentially methylated gene
Differentially methylated gene
| Probe | Chromosome | Position | Nearest gene | P (dis) | Beta (dis) | FDR (dis) | Study |
|---|---|---|---|---|---|---|---|
| cg10904715 | 13 | 25115837 | PARP4 | 3.29E-8 | -0.013 | 9.87E-6 | DMG:Jaffe_2016 |
Section II. Transcriptome annotation
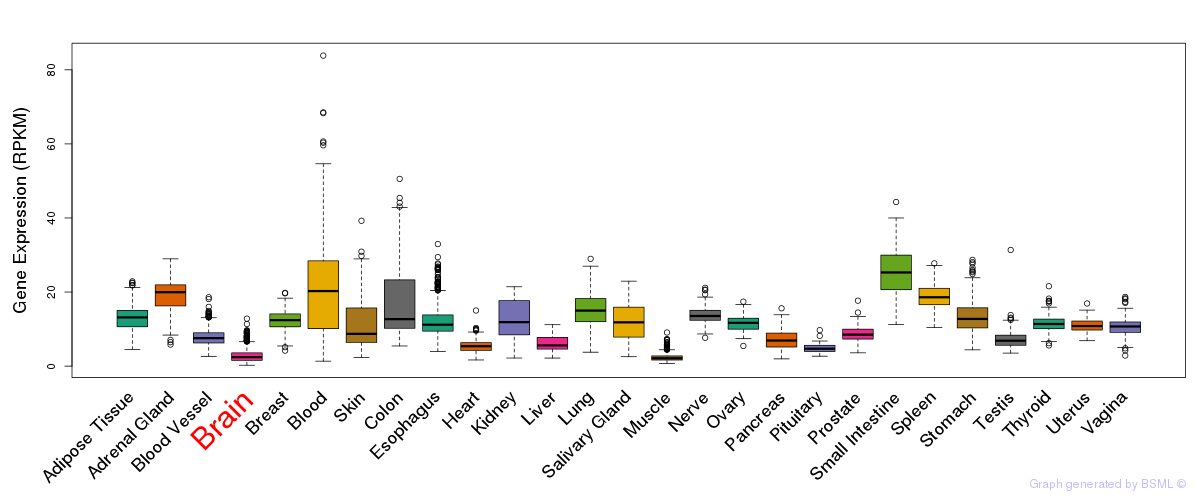
General gene expression (GTEx)
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
Top co-expressed genes in brain regions
| Top 10 positively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| KCNC2 | 0.82 | 0.76 |
| RASSF5 | 0.81 | 0.84 |
| ANKRD34C | 0.81 | 0.85 |
| NPTX2 | 0.80 | 0.85 |
| TRPM2 | 0.80 | 0.84 |
| SSTR3 | 0.80 | 0.88 |
| PTHLH | 0.80 | 0.84 |
| GPR123 | 0.80 | 0.78 |
| ANXA6 | 0.80 | 0.86 |
| SGPP2 | 0.79 | 0.82 |
| Top 10 negatively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| RBMX2 | -0.44 | -0.63 |
| FADS2 | -0.43 | -0.49 |
| TUBB2B | -0.43 | -0.59 |
| TRAF4 | -0.43 | -0.61 |
| RPL23A | -0.43 | -0.60 |
| GTF3C6 | -0.42 | -0.50 |
| KIAA1949 | -0.42 | -0.48 |
| CARHSP1 | -0.42 | -0.64 |
| PDE9A | -0.41 | -0.58 |
| SEMA4B | -0.41 | -0.48 |
Section III. Gene Ontology annotation
| Molecular function | GO term | Evidence | Neuro keywords | PubMed ID |
|---|---|---|---|---|
| GO:0003950 | NAD+ ADP-ribosyltransferase activity | NAS | 10644454 | |
| GO:0003677 | DNA binding | TAS | 10100603 | |
| GO:0016757 | transferase activity, transferring glycosyl groups | IEA | - | |
| GO:0019899 | enzyme binding | IDA | 15169895 | |
| Biological process | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0006464 | protein modification process | TAS | 10100603 | |
| GO:0006471 | protein amino acid ADP-ribosylation | NAS | 10644454 | |
| GO:0008219 | cell death | IMP | 12140175 | |
| GO:0006281 | DNA repair | NAS | 12101391 | |
| GO:0006810 | transport | NAS | 11855821 | |
| GO:0006954 | inflammatory response | IMP | 12123754 | |
| GO:0042493 | response to drug | NAS | 11291045 | |
| Cellular component | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0005622 | intracellular | IEA | - | |
| GO:0005634 | nucleus | NAS | 10644454 | |
| GO:0005737 | cytoplasm | IEA | - | |
| GO:0030529 | ribonucleoprotein complex | NAS | 11479319 |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| KEGG BASE EXCISION REPAIR | 35 | 22 | All SZGR 2.0 genes in this pathway |
| CHEMNITZ RESPONSE TO PROSTAGLANDIN E2 DN | 391 | 222 | All SZGR 2.0 genes in this pathway |
| CHARAFE BREAST CANCER LUMINAL VS BASAL DN | 455 | 304 | All SZGR 2.0 genes in this pathway |
| OSMAN BLADDER CANCER UP | 404 | 246 | All SZGR 2.0 genes in this pathway |
| JAERVINEN AMPLIFIED IN LARYNGEAL CANCER | 40 | 24 | All SZGR 2.0 genes in this pathway |
| AIYAR COBRA1 TARGETS UP | 39 | 25 | All SZGR 2.0 genes in this pathway |
| MEINHOLD OVARIAN CANCER LOW GRADE UP | 19 | 15 | All SZGR 2.0 genes in this pathway |
| GRUETZMANN PANCREATIC CANCER UP | 358 | 245 | All SZGR 2.0 genes in this pathway |
| SPIELMAN LYMPHOBLAST EUROPEAN VS ASIAN DN | 584 | 395 | All SZGR 2.0 genes in this pathway |
| SUNG METASTASIS STROMA DN | 54 | 34 | All SZGR 2.0 genes in this pathway |
| KAUFFMANN DNA REPAIR GENES | 230 | 137 | All SZGR 2.0 genes in this pathway |
| SHEPARD BMYB MORPHOLINO UP | 205 | 126 | All SZGR 2.0 genes in this pathway |
| DER IFN BETA RESPONSE UP | 102 | 67 | All SZGR 2.0 genes in this pathway |
| FAELT B CLL WITH VH REARRANGEMENTS DN | 48 | 26 | All SZGR 2.0 genes in this pathway |
| BROWNE HCMV INFECTION 10HR DN | 56 | 37 | All SZGR 2.0 genes in this pathway |
| CUI TCF21 TARGETS 2 DN | 830 | 547 | All SZGR 2.0 genes in this pathway |
| BROWNE HCMV INFECTION 14HR DN | 298 | 200 | All SZGR 2.0 genes in this pathway |
| CUI TCF21 TARGETS DN | 31 | 23 | All SZGR 2.0 genes in this pathway |
| WANG CISPLATIN RESPONSE AND XPC DN | 228 | 146 | All SZGR 2.0 genes in this pathway |
| DEBIASI APOPTOSIS BY REOVIRUS INFECTION DN | 287 | 208 | All SZGR 2.0 genes in this pathway |
| BROWNE HCMV INFECTION 6HR DN | 160 | 101 | All SZGR 2.0 genes in this pathway |
| CREIGHTON ENDOCRINE THERAPY RESISTANCE 4 | 307 | 185 | All SZGR 2.0 genes in this pathway |
| MASSARWEH TAMOXIFEN RESISTANCE DN | 258 | 160 | All SZGR 2.0 genes in this pathway |
| ACEVEDO LIVER TUMOR VS NORMAL ADJACENT TISSUE UP | 863 | 514 | All SZGR 2.0 genes in this pathway |
| GRADE COLON CANCER UP | 871 | 505 | All SZGR 2.0 genes in this pathway |
| AGUIRRE PANCREATIC CANCER COPY NUMBER UP | 298 | 174 | All SZGR 2.0 genes in this pathway |
| MILI PSEUDOPODIA HAPTOTAXIS UP | 518 | 299 | All SZGR 2.0 genes in this pathway |
| YOSHIOKA LIVER CANCER EARLY RECURRENCE DN | 65 | 31 | All SZGR 2.0 genes in this pathway |
| WANG METASTASIS OF BREAST CANCER | 15 | 8 | All SZGR 2.0 genes in this pathway |
| KOINUMA TARGETS OF SMAD2 OR SMAD3 | 824 | 528 | All SZGR 2.0 genes in this pathway |