Gene Page: ADRA1B
Summary ?
| GeneID | 147 |
| Symbol | ADRA1B |
| Synonyms | ADRA1|ALPHA1BAR |
| Description | adrenoceptor alpha 1B |
| Reference | MIM:104220|HGNC:HGNC:278|HPRD:00080| |
| Gene type | protein-coding |
| Map location | 5q33.3 |
| Pascal p-value | 0.507 |
| Fetal beta | -0.89 |
| eGene | Meta |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| ADT:Sun_2012 | Systematic Investigation of Antipsychotic Drugs and Their Targets | A total of 382 drug-target associations involving 43 antipsychotic drugs and 49 target genes. | |
| CV:GWASdb | Genome-wide Association Studies | GWASdb records for schizophrenia | |
| CV:PGCnp | Genome-wide Association Study | GWAS | |
| GSMA_I | Genome scan meta-analysis | Psr: 0.0032 | |
| GSMA_IIE | Genome scan meta-analysis (European-ancestry samples) | Psr: 0.01718 | |
| GSMA_IIA | Genome scan meta-analysis (All samples) | Psr: 0.00459 | |
| Network | Shortest path distance of core genes in the Human protein-protein interaction network | Contribution to shortest path in PPI network: 0.0483 |
Section I. Genetics and epigenetics annotation
Section II. Transcriptome annotation
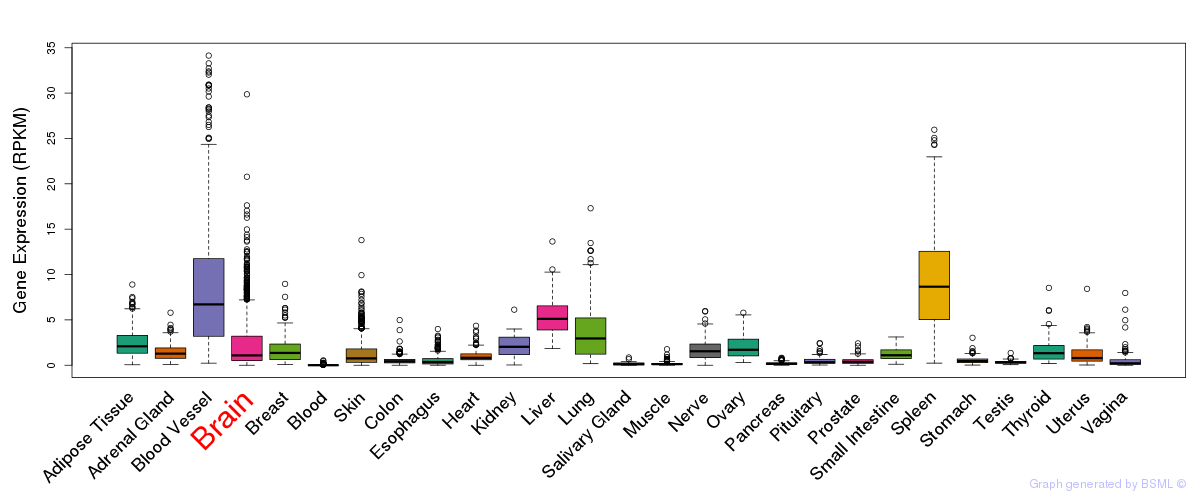
General gene expression (GTEx)
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
Top co-expressed genes in brain regions
| Top 10 positively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| MN1 | 0.91 | 0.89 |
| SOBP | 0.90 | 0.94 |
| MPPED1 | 0.89 | 0.92 |
| SCUBE1 | 0.88 | 0.81 |
| SLA | 0.88 | 0.88 |
| TTC28 | 0.88 | 0.87 |
| SATB2 | 0.88 | 0.90 |
| NHSL1 | 0.87 | 0.70 |
| SLC22A23 | 0.87 | 0.87 |
| MYCN | 0.87 | 0.89 |
| Top 10 negatively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| SERPINB6 | -0.60 | -0.76 |
| ACOT13 | -0.57 | -0.67 |
| HEPN1 | -0.57 | -0.74 |
| AIFM3 | -0.57 | -0.74 |
| HSD17B14 | -0.57 | -0.77 |
| C5orf53 | -0.57 | -0.72 |
| HLA-F | -0.56 | -0.73 |
| ALDOC | -0.56 | -0.67 |
| CLU | -0.56 | -0.68 |
| TSC22D4 | -0.56 | -0.76 |
Section III. Gene Ontology annotation
| Molecular function | GO term | Evidence | Neuro keywords | PubMed ID |
|---|---|---|---|---|
| GO:0004872 | receptor activity | IEA | - | |
| GO:0004937 | alpha1-adrenergic receptor activity | TAS | 1328250 | |
| GO:0004935 | adrenoceptor activity | IEA | - | |
| Biological process | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0001987 | vasoconstriction of artery involved in baroreceptor response to lowering of systemic arterial blood pressure | IEA | - | |
| GO:0001996 | positive regulation of heart rate by epinephrine-norepinephrine | IEA | - | |
| GO:0001997 | positive regulation of the force of heart contraction by epinephrine-norepinephrine | IEA | - | |
| GO:0001975 | response to amphetamine | IEA | - | |
| GO:0001974 | blood vessel remodeling | IEA | - | |
| GO:0007188 | G-protein signaling, coupled to cAMP nucleotide second messenger | TAS | 10820200 | |
| GO:0007186 | G-protein coupled receptor protein signaling pathway | TAS | 2154750 | |
| GO:0016049 | cell growth | IEA | - | |
| GO:0007267 | cell-cell signaling | TAS | 1328250 | |
| GO:0007243 | protein kinase cascade | TAS | 10820200 | |
| GO:0007165 | signal transduction | IEA | - | |
| GO:0008283 | cell proliferation | TAS | 10820200 | |
| GO:0008542 | visual learning | IEA | - | |
| GO:0007512 | adult heart development | IEA | - | |
| GO:0007626 | locomotory behavior | IEA | - | |
| GO:0007275 | multicellular organismal development | TAS | 10820200 | |
| GO:0042593 | glucose homeostasis | IEA | - | |
| GO:0035265 | organ growth | IEA | - | |
| GO:0043278 | response to morphine | IEA | - | |
| GO:0048148 | behavioral response to cocaine | IEA | - | |
| GO:0045819 | positive regulation of glycogen catabolic process | IEA | - | |
| GO:0045818 | negative regulation of glycogen catabolic process | IEA | - | |
| Cellular component | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0000299 | integral to membrane of membrane fraction | IEA | - | |
| GO:0005886 | plasma membrane | IEA | - | |
| GO:0005887 | integral to plasma membrane | TAS | 1328250 |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| KEGG CALCIUM SIGNALING PATHWAY | 178 | 134 | All SZGR 2.0 genes in this pathway |
| KEGG NEUROACTIVE LIGAND RECEPTOR INTERACTION | 272 | 195 | All SZGR 2.0 genes in this pathway |
| KEGG VASCULAR SMOOTH MUSCLE CONTRACTION | 115 | 81 | All SZGR 2.0 genes in this pathway |
| PID LYSOPHOSPHOLIPID PATHWAY | 66 | 53 | All SZGR 2.0 genes in this pathway |
| REACTOME GASTRIN CREB SIGNALLING PATHWAY VIA PKC AND MAPK | 205 | 136 | All SZGR 2.0 genes in this pathway |
| REACTOME SIGNALING BY GPCR | 920 | 449 | All SZGR 2.0 genes in this pathway |
| REACTOME CLASS A1 RHODOPSIN LIKE RECEPTORS | 305 | 177 | All SZGR 2.0 genes in this pathway |
| REACTOME AMINE LIGAND BINDING RECEPTORS | 38 | 33 | All SZGR 2.0 genes in this pathway |
| REACTOME G ALPHA Q SIGNALLING EVENTS | 184 | 116 | All SZGR 2.0 genes in this pathway |
| REACTOME G ALPHA1213 SIGNALLING EVENTS | 74 | 56 | All SZGR 2.0 genes in this pathway |
| REACTOME GPCR DOWNSTREAM SIGNALING | 805 | 368 | All SZGR 2.0 genes in this pathway |
| REACTOME GPCR LIGAND BINDING | 408 | 246 | All SZGR 2.0 genes in this pathway |
| PETRETTO HEART MASS QTL CIS UP | 28 | 15 | All SZGR 2.0 genes in this pathway |
| BANDRES RESPONSE TO CARMUSTIN MGMT 24HR DN | 33 | 21 | All SZGR 2.0 genes in this pathway |
| BANDRES RESPONSE TO CARMUSTIN WITHOUT MGMT 48HR DN | 32 | 25 | All SZGR 2.0 genes in this pathway |
| SEMENZA HIF1 TARGETS | 36 | 32 | All SZGR 2.0 genes in this pathway |
| BANDRES RESPONSE TO CARMUSTIN MGMT 48HR DN | 161 | 105 | All SZGR 2.0 genes in this pathway |
| ACEVEDO LIVER TUMOR VS NORMAL ADJACENT TISSUE DN | 274 | 165 | All SZGR 2.0 genes in this pathway |
| MEISSNER BRAIN HCP WITH H3K4ME3 AND H3K27ME3 | 1069 | 729 | All SZGR 2.0 genes in this pathway |
| MEISSNER NPC HCP WITH H3K4ME2 | 491 | 319 | All SZGR 2.0 genes in this pathway |
| DANG MYC TARGETS UP | 143 | 100 | All SZGR 2.0 genes in this pathway |
| DANG BOUND BY MYC | 1103 | 714 | All SZGR 2.0 genes in this pathway |
| OHGUCHI LIVER HNF4A TARGETS DN | 149 | 85 | All SZGR 2.0 genes in this pathway |
| SERVITJA LIVER HNF1A TARGETS DN | 157 | 105 | All SZGR 2.0 genes in this pathway |
| FORTSCHEGGER PHF8 TARGETS DN | 784 | 464 | All SZGR 2.0 genes in this pathway |
| PEDRIOLI MIR31 TARGETS UP | 221 | 120 | All SZGR 2.0 genes in this pathway |
| ZWANG TRANSIENTLY UP BY 2ND EGF PULSE ONLY | 1725 | 838 | All SZGR 2.0 genes in this pathway |
Section VI. microRNA annotation
| miRNA family | Target position | miRNA ID | miRNA seq | ||
|---|---|---|---|---|---|
| UTR start | UTR end | Match method | |||
| miR-22 | 141 | 147 | m8 | hsa-miR-22brain | AAGCUGCCAGUUGAAGAACUGU |
- SZ: miRNAs which differentially expressed in brain cortex of schizophrenia patients comparing with control samples using microarray. Click here to see the list of SZ related miRNAs.
- Brain: miRNAs which are expressed in brain based on miRNA microarray expression studies. Click here to see the list of brain related miRNAs.