Gene Page: AES
Summary ?
| GeneID | 166 |
| Symbol | AES |
| Synonyms | AES-1|AES-2|ESP1|GRG|GRG5|Grg-5|TLE5 |
| Description | amino-terminal enhancer of split |
| Reference | MIM:600188|HGNC:HGNC:307|Ensembl:ENSG00000104964|HPRD:02556|Vega:OTTHUMG00000180567 |
| Gene type | protein-coding |
| Map location | 19p13.3 |
| Pascal p-value | 0.422 |
| Sherlock p-value | 0.465 |
| Fetal beta | -0.748 |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CV:PGCnp | Genome-wide Association Study | GWAS | |
| PMID:cooccur | High-throughput literature-search | Systematic search in PubMed for genes co-occurring with SCZ keywords. A total of 3027 genes were included. | |
| Literature | High-throughput literature-search | Co-occurance with Schizophrenia keywords: schizophrenia,schizophrenic,schizophrenias | Click to show details |
Section I. Genetics and epigenetics annotation
Section II. Transcriptome annotation
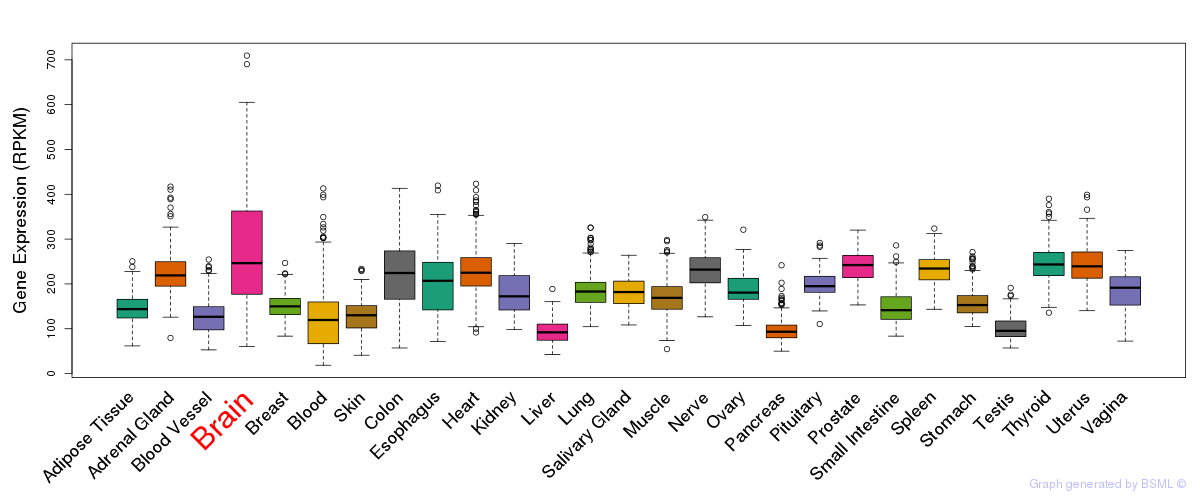
General gene expression (GTEx)
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
No co-expressed genes in brain regions
Section IV. Protein-protein interaction annotation
| Interactors | Aliases B | Official full name B | Experimental | Source | PubMed ID |
|---|---|---|---|---|---|
| AES | AES-1 | AES-2 | ESP1 | GRG | GRG5 | TLE5 | amino-terminal enhancer of split | AES interacts with AES. | BIND | 10660609 |
| AES | AES-1 | AES-2 | ESP1 | GRG | GRG5 | TLE5 | amino-terminal enhancer of split | Two-hybrid | BioGRID | 11266540 |
| AR | AIS | DHTR | HUMARA | KD | NR3C4 | SBMA | SMAX1 | TFM | androgen receptor | - | HPRD,BioGRID | 11416139 |
| GTF2E1 | FE | TF2E1 | TFIIE-A | general transcription factor IIE, polypeptide 1, alpha 56kDa | - | HPRD,BioGRID | 11416139 |
| HMGB1 | DKFZp686A04236 | HMG1 | HMG3 | SBP-1 | high-mobility group box 1 | - | HPRD,BioGRID | 11748221 |
| HNF1A | HNF1 | LFB1 | MODY3 | TCF1 | HNF1 homeobox A | Two-hybrid | BioGRID | 11266540 |
| HNRNPAB | ABBP1 | FLJ40338 | HNRPAB | heterogeneous nuclear ribonucleoprotein A/B | Affinity Capture-MS | BioGRID | 17353931 |
| MED31 | 3110004H13Rik | CGI-125 | FLJ27436 | FLJ36714 | Soh1 | mediator complex subunit 31 | Two-hybrid | BioGRID | 16169070 |
| PTRH2 | BIT1 | CGI-147 | FLJ32471 | PTH2 | peptidyl-tRNA hydrolase 2 | Bit1 interacts with AES. | BIND | 15006356 |
| PTRH2 | BIT1 | CGI-147 | FLJ32471 | PTH2 | peptidyl-tRNA hydrolase 2 | - | HPRD,BioGRID | 15006356 |
| RELA | MGC131774 | NFKB3 | p65 | v-rel reticuloendotheliosis viral oncogene homolog A (avian) | AES interacts with p65. | BIND | 10660609 |
| RELA | MGC131774 | NFKB3 | p65 | v-rel reticuloendotheliosis viral oncogene homolog A (avian) | - | HPRD,BioGRID | 10660609 |
| RUNX1 | AML1 | AML1-EVI-1 | AMLCR1 | CBFA2 | EVI-1 | PEBP2aB | runt-related transcription factor 1 | Two-hybrid | BioGRID | 10825294 |
| SH3GL3 | CNSA3 | EEN-2B-L3 | EEN-B2 | HsT19371 | SH3D2C | SH3P13 | SH3-domain GRB2-like 3 | Two-hybrid | BioGRID | 16169070 |
| SIX1 | BOS3 | DFNA23 | TIP39 | SIX homeobox 1 | SIX1 interacts with AES. | BIND | 12441302 |
| SIX2 | - | SIX homeobox 2 | SIX2 interacts with AES. This interaction was modeled on a demonstrated interaction between mouse Six2 and human AES. | BIND | 12441302 |
| SIX3 | HPE2 | SIX homeobox 3 | Reconstituted Complex Two-hybrid | BioGRID | 12441302 |
| SIX3 | HPE2 | SIX homeobox 3 | - | HPRD | 12050133 |
| SIX3 | HPE2 | SIX homeobox 3 | SIX3 interacts with AES. | BIND | 12441302 |
| SIX6 | MCOPCT2 | OPTX2 | Six9 | SIX homeobox 6 | Reconstituted Complex Two-hybrid | BioGRID | 12441302 |
| SIX6 | MCOPCT2 | OPTX2 | Six9 | SIX homeobox 6 | SIX6 interacts with AES. | BIND | 12441302 |
| STYXL1 | DUSP24 | MK-STYX | serine/threonine/tyrosine interacting-like 1 | Affinity Capture-MS | BioGRID | 17353931 |
| TLE1 | ESG | ESG1 | GRG1 | transducin-like enhancer of split 1 (E(sp1) homolog, Drosophila) | Affinity Capture-MS | BioGRID | 17353931 |
| TLE3 | ESG | ESG3 | FLJ39460 | GRG3 | HsT18976 | KIAA1547 | transducin-like enhancer of split 3 (E(sp1) homolog, Drosophila) | Affinity Capture-MS | BioGRID | 17353931 |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| WNT SIGNALING | 89 | 71 | All SZGR 2.0 genes in this pathway |
| PID PS1 PATHWAY | 46 | 39 | All SZGR 2.0 genes in this pathway |
| PID BETA CATENIN NUC PATHWAY | 80 | 60 | All SZGR 2.0 genes in this pathway |
| NAKAMURA TUMOR ZONE PERIPHERAL VS CENTRAL DN | 634 | 384 | All SZGR 2.0 genes in this pathway |
| BARRIER COLON CANCER RECURRENCE DN | 20 | 16 | All SZGR 2.0 genes in this pathway |
| BERENJENO TRANSFORMED BY RHOA DN | 394 | 258 | All SZGR 2.0 genes in this pathway |
| DACOSTA UV RESPONSE VIA ERCC3 TTD UP | 64 | 39 | All SZGR 2.0 genes in this pathway |
| HUMMERICH SKIN CANCER PROGRESSION DN | 100 | 64 | All SZGR 2.0 genes in this pathway |
| SPIELMAN LYMPHOBLAST EUROPEAN VS ASIAN UP | 479 | 299 | All SZGR 2.0 genes in this pathway |
| NUYTTEN NIPP1 TARGETS DN | 848 | 527 | All SZGR 2.0 genes in this pathway |
| KLEIN PRIMARY EFFUSION LYMPHOMA DN | 58 | 42 | All SZGR 2.0 genes in this pathway |
| LIN APC TARGETS | 77 | 55 | All SZGR 2.0 genes in this pathway |
| POMEROY MEDULLOBLASTOMA PROGNOSIS UP | 47 | 30 | All SZGR 2.0 genes in this pathway |
| PENG RAPAMYCIN RESPONSE UP | 203 | 130 | All SZGR 2.0 genes in this pathway |
| LENAOUR DENDRITIC CELL MATURATION UP | 114 | 84 | All SZGR 2.0 genes in this pathway |
| BLALOCK ALZHEIMERS DISEASE DN | 1237 | 837 | All SZGR 2.0 genes in this pathway |
| BURTON ADIPOGENESIS PEAK AT 0HR | 63 | 48 | All SZGR 2.0 genes in this pathway |
| XU GH1 AUTOCRINE TARGETS UP | 268 | 157 | All SZGR 2.0 genes in this pathway |
| DOUGLAS BMI1 TARGETS UP | 566 | 371 | All SZGR 2.0 genes in this pathway |
| STEARMAN LUNG CANCER EARLY VS LATE DN | 61 | 39 | All SZGR 2.0 genes in this pathway |
| MCCABE BOUND BY HOXC6 | 469 | 239 | All SZGR 2.0 genes in this pathway |
| MOOTHA PGC | 420 | 269 | All SZGR 2.0 genes in this pathway |
| YOSHIMURA MAPK8 TARGETS UP | 1305 | 895 | All SZGR 2.0 genes in this pathway |
| MUELLER COMMON TARGETS OF AML FUSIONS UP | 14 | 10 | All SZGR 2.0 genes in this pathway |
| SWEET LUNG CANCER KRAS UP | 491 | 316 | All SZGR 2.0 genes in this pathway |
| HSIAO HOUSEKEEPING GENES | 389 | 245 | All SZGR 2.0 genes in this pathway |
| JISON SICKLE CELL DISEASE DN | 181 | 97 | All SZGR 2.0 genes in this pathway |
| YAGI AML WITH T 8 21 TRANSLOCATION | 368 | 247 | All SZGR 2.0 genes in this pathway |
| KARLSSON TGFB1 TARGETS DN | 207 | 139 | All SZGR 2.0 genes in this pathway |
| QI HYPOXIA TARGETS OF HIF1A AND FOXA2 | 37 | 27 | All SZGR 2.0 genes in this pathway |
| JUBAN TARGETS OF SPI1 AND FLI1 UP | 115 | 73 | All SZGR 2.0 genes in this pathway |
| DELACROIX RAR BOUND ES | 462 | 273 | All SZGR 2.0 genes in this pathway |
| ALFANO MYC TARGETS | 239 | 156 | All SZGR 2.0 genes in this pathway |
| KUMAR PATHOGEN LOAD BY MACROPHAGES | 275 | 155 | All SZGR 2.0 genes in this pathway |
| HOLLEMAN PREDNISOLONE RESISTANCE B ALL UP | 22 | 16 | All SZGR 2.0 genes in this pathway |
| HOLLEMAN PREDNISOLONE RESISTANCE ALL UP | 19 | 15 | All SZGR 2.0 genes in this pathway |