Gene Page: DFFA
Summary ?
| GeneID | 1676 |
| Symbol | DFFA |
| Synonyms | DFF-45|DFF1|ICAD |
| Description | DNA fragmentation factor subunit alpha |
| Reference | MIM:601882|HGNC:HGNC:2772|HPRD:03531| |
| Gene type | protein-coding |
| Map location | 1p36.3-p36.2 |
| Pascal p-value | 0.465 |
| Sherlock p-value | 0.526 |
| Fetal beta | -0.276 |
| DMG | 2 (# studies) |
| eGene | Frontal Cortex BA9 Myers' cis & trans |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| DMG:Jaffe_2016 | Genome-wide DNA methylation analysis | This dataset includes 2,104 probes/CpGs associated with SZ patients (n=108) compared to 136 controls at Bonferroni-adjusted P < 0.05. | 2 |
| DMG:Wockner_2014 | Genome-wide DNA methylation analysis | This dataset includes 4641 differentially methylated probes corresponding to 2929 unique genes between schizophrenia patients (n=24) and controls (n=24). | 2 |
| Network | Shortest path distance of core genes in the Human protein-protein interaction network | Contribution to shortest path in PPI network: 0.0069 |
Section I. Genetics and epigenetics annotation
 Differentially methylated gene
Differentially methylated gene
| Probe | Chromosome | Position | Nearest gene | P (dis) | Beta (dis) | FDR (dis) | Study |
|---|---|---|---|---|---|---|---|
| cg08029392 | 1 | 10521539 | DFFA | 1.86E-4 | 0.397 | 0.034 | DMG:Wockner_2014 |
| cg15452426 | 1 | 10532230 | DFFA | 9.46E-8 | -0.007 | 2.13E-5 | DMG:Jaffe_2016 |
 eQTL annotation
eQTL annotation
| SNP ID | Chromosome | Position | eGene | Gene Entrez ID | pvalue | qvalue | TSS distance | eQTL type |
|---|---|---|---|---|---|---|---|---|
| rs7742891 | chr6 | 49525359 | DFFA | 1676 | 0.11 | trans | ||
| rs911165 | chr20 | 4544778 | DFFA | 1676 | 0.06 | trans |
Section II. Transcriptome annotation
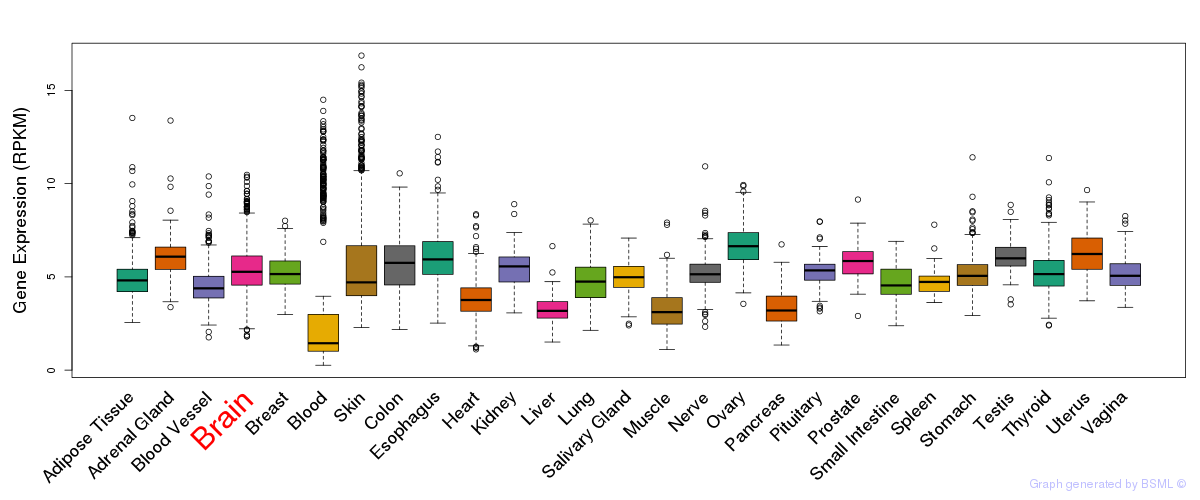
General gene expression (GTEx)
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
No co-expressed genes in brain regions
Section III. Gene Ontology annotation
| Molecular function | GO term | Evidence | Neuro keywords | PubMed ID |
|---|---|---|---|---|
| GO:0004537 | caspase-activated deoxyribonuclease activity | TAS | 9108473 | |
| GO:0042802 | identical protein binding | IPI | 17353931 | |
| Biological process | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0006309 | DNA fragmentation during apoptosis | IDA | 15572351 | |
| GO:0007242 | intracellular signaling cascade | TAS | 9108473 | |
| GO:0006917 | induction of apoptosis | IEA | - | |
| GO:0043066 | negative regulation of apoptosis | IDA | 9564035 | |
| Cellular component | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0005829 | cytosol | TAS | 9108473 | |
| GO:0005622 | intracellular | IEA | - | |
| GO:0005634 | nucleus | IDA | 15572351 | |
| GO:0005654 | nucleoplasm | EXP | 9108473 | |
| GO:0005737 | cytoplasm | IEA | - |
Section IV. Protein-protein interaction annotation
| Interactors | Aliases B | Official full name B | Experimental | Source | PubMed ID |
|---|---|---|---|---|---|
| BAK1 | BAK | BAK-LIKE | BCL2L7 | CDN1 | MGC117255 | MGC3887 | BCL2-antagonist/killer 1 | Affinity Capture-MS | BioGRID | 17353931 |
| CASP3 | CPP32 | CPP32B | SCA-1 | caspase 3, apoptosis-related cysteine peptidase | - | HPRD | 11752060 |
| CIDEB | - | cell death-inducing DFFA-like effector b | - | HPRD,BioGRID | 10619428 |
| DFFB | CAD | CPAN | DFF-40 | DFF2 | DFF40 | DNA fragmentation factor, 40kDa, beta polypeptide (caspase-activated DNase) | - | HPRD | 9422506 |10409614 |10527860 |10527861 |10932250 |11371636 |
| DFFB | CAD | CPAN | DFF-40 | DFF2 | DFF40 | DNA fragmentation factor, 40kDa, beta polypeptide (caspase-activated DNase) | Affinity Capture-MS Affinity Capture-Western | BioGRID | 10527860 |17353931 |
| EWSR1 | EWS | Ewing sarcoma breakpoint region 1 | Two-hybrid | BioGRID | 16189514 |
| HERC1 | p532 | p619 | hect (homologous to the E6-AP (UBE3A) carboxyl terminus) domain and RCC1 (CHC1)-like domain (RLD) 1 | Affinity Capture-MS | BioGRID | 17353931 |
| MCC | DKFZp762O1615 | FLJ38893 | FLJ46755 | MCC1 | mutated in colorectal cancers | Affinity Capture-MS | BioGRID | 17353931 |
| NAP1L5 | DRLM | nucleosome assembly protein 1-like 5 | Affinity Capture-MS | BioGRID | 17353931 |
| NECAB2 | EFCBP2 | N-terminal EF-hand calcium binding protein 2 | Two-hybrid | BioGRID | 16189514 |
| TMBIM4 | CGI-119 | GAAP | S1R | ZPRO | transmembrane BAX inhibitor motif containing 4 | Affinity Capture-MS | BioGRID | 17353931 |
| USP15 | KIAA0529 | MGC131982 | MGC149838 | MGC74854 | UNPH4 | ubiquitin specific peptidase 15 | Affinity Capture-MS | BioGRID | 17353931 |
| YWHAG | 14-3-3GAMMA | tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein, gamma polypeptide | - | HPRD | 15324660 |
| YWHAZ | KCIP-1 | MGC111427 | MGC126532 | MGC138156 | tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein, zeta polypeptide | Affinity Capture-MS | BioGRID | 17353931 |