Gene Page: DNM1
Summary ?
| GeneID | 1759 |
| Symbol | DNM1 |
| Synonyms | DNM|EIEE31 |
| Description | dynamin 1 |
| Reference | MIM:602377|HGNC:HGNC:2972|Ensembl:ENSG00000106976|HPRD:03851|Vega:OTTHUMG00000020733 |
| Gene type | protein-coding |
| Map location | 9q34 |
| Pascal p-value | 0.058 |
| Sherlock p-value | 0.923 |
| Fetal beta | -2.426 |
| Support | ENDOCYTOSIS G2Cdb.human_BAYES-COLLINS-HUMAN-PSD-CONSENSUS G2Cdb.human_BAYES-COLLINS-HUMAN-PSD-FULL G2Cdb.human_BAYES-COLLINS-MOUSE-PSD-CONSENSUS G2Cdb.human_clathrin G2Cdb.human_mGluR5 G2Cdb.human_Synaptosome G2Cdb.humanNRC G2Cdb.humanPSD G2Cdb.humanPSP Darnell FMRP targets Potential synaptic genes |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CV:PGCnp | Genome-wide Association Study | GWAS | |
| DMG:Wockner_2014 | Genome-wide DNA methylation analysis | This dataset includes 4641 differentially methylated probes corresponding to 2929 unique genes between schizophrenia patients (n=24) and controls (n=24). | 1 |
| PMID:cooccur | High-throughput literature-search | Systematic search in PubMed for genes co-occurring with SCZ keywords. A total of 3027 genes were included. | |
| Literature | High-throughput literature-search | Co-occurance with Schizophrenia keywords: schizophrenia,schizophrenias | Click to show details |
| Network | Shortest path distance of core genes in the Human protein-protein interaction network | Contribution to shortest path in PPI network: 0.3528 |
Section I. Genetics and epigenetics annotation
 Differentially methylated gene
Differentially methylated gene
| Probe | Chromosome | Position | Nearest gene | P (dis) | Beta (dis) | FDR (dis) | Study |
|---|---|---|---|---|---|---|---|
| cg14409940 | 9 | 130967820 | CIZ1;DNM1 | 2.485E-4 | 0.432 | 0.037 | DMG:Wockner_2014 |
Section II. Transcriptome annotation
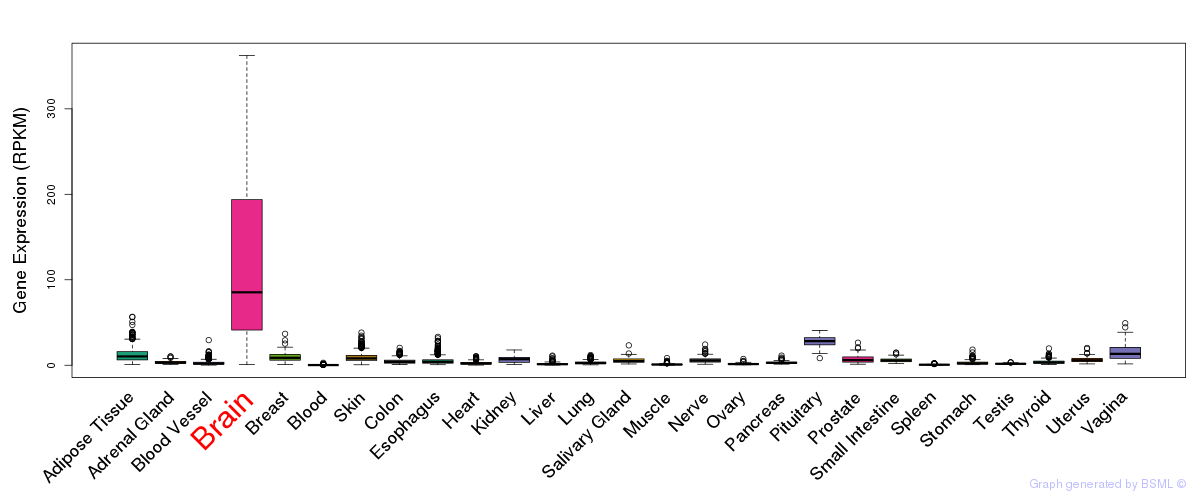
General gene expression (GTEx)
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
No co-expressed genes in brain regions
Section III. Gene Ontology annotation
| Molecular function | GO term | Evidence | Neuro keywords | PubMed ID |
|---|---|---|---|---|
| GO:0000166 | nucleotide binding | IEA | - | |
| GO:0003774 | motor activity | IEA | - | |
| GO:0003924 | GTPase activity | IEA | - | |
| GO:0003924 | GTPase activity | TAS | 10944110 | |
| GO:0005515 | protein binding | ISS | - | |
| GO:0005525 | GTP binding | IEA | - | |
| GO:0016787 | hydrolase activity | IEA | - | |
| Biological process | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0006898 | receptor-mediated endocytosis | TAS | 8101525 | |
| Cellular component | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0005856 | cytoskeleton | IEA | - | |
| GO:0005874 | microtubule | IEA | - | |
| GO:0005737 | cytoplasm | IEA | - |
Section IV. Protein-protein interaction annotation
| Interactors | Aliases B | Official full name B | Experimental | Source | PubMed ID |
|---|---|---|---|---|---|
| AMPH | AMPH1 | amphiphysin | Affinity Capture-Western Reconstituted Complex | BioGRID | 9148966 |9280305 |9341169 |9348539 |11877424 |
| AMPH | AMPH1 | amphiphysin | - | HPRD | 9148966 |9348539 |9694653 |11877424|10899172 |
| AP2A2 | ADTAB | CLAPA2 | HIP9 | HYPJ | adaptor-related protein complex 2, alpha 2 subunit | - | HPRD | 10430869 |
| ASB13 | FLJ13134 | MGC19879 | ankyrin repeat and SOCS box-containing 13 | Two-hybrid | BioGRID | 16169070 |
| BIN1 | AMPH2 | AMPHL | DKFZp547F068 | MGC10367 | SH3P9 | bridging integrator 1 | - | HPRD,BioGRID | 9195986 |9280305 |
| CABIN1 | CAIN | KIAA0330 | PPP3IN | calcineurin binding protein 1 | - | HPRD,BioGRID | 10931822 |
| CLIC3 | - | chloride intracellular channel 3 | - | HPRD | 11563969 |
| CLIC4 | CLIC4L | DKFZp566G223 | FLJ38640 | H1 | MTCLIC | huH1 | p64H1 | chloride intracellular channel 4 | - | HPRD | 11563969 |
| CLINT1 | CLINT | ENTH | EPN4 | EPNR | EPSINR | FLJ46753 | KIAA0171 | clathrin interactor 1 | Reconstituted Complex | BioGRID | 12429846 |
| CTTN | EMS1 | FLJ34459 | cortactin | - | HPRD | 12419186 |
| DNM1 | DNM | dynamin 1 | - | HPRD,BioGRID | 7634088 |
| DNMBP | KIAA1010 | TUBA | dynamin binding protein | - | HPRD,BioGRID | 14506234 |
| DYRK1A | DYRK | DYRK1 | HP86 | MNB | MNBH | dual-specificity tyrosine-(Y)-phosphorylation regulated kinase 1A | Reconstituted Complex | BioGRID | 11877424 |
| EPS15 | AF-1P | AF1P | MLLT5 | epidermal growth factor receptor pathway substrate 15 | - | HPRD | 11483962 |
| FER | TYK3 | fer (fps/fes related) tyrosine kinase | Co-fractionation | BioGRID | 9722593 |
| FNBP1 | FBP17 | KIAA0554 | MGC126804 | formin binding protein 1 | - | HPRD,BioGRID | 15252009 |
| GNB2L1 | Gnb2-rs1 | H12.3 | HLC-7 | PIG21 | RACK1 | guanine nucleotide binding protein (G protein), beta polypeptide 2-like 1 | - | HPRD | 12359736 |
| GRB2 | ASH | EGFRBP-GRB2 | Grb3-3 | MST084 | MSTP084 | growth factor receptor-bound protein 2 | Affinity Capture-Western Far Western Reconstituted Complex | BioGRID | 8119878 |9009162 |
| GRB2 | ASH | EGFRBP-GRB2 | Grb3-3 | MST084 | MSTP084 | growth factor receptor-bound protein 2 | - | HPRD | 8119878 |11877424 |
| GRIN1 | NMDA1 | NMDAR1 | NR1 | glutamate receptor, ionotropic, N-methyl D-aspartate 1 | - | HPRD | 10862698 |
| GRIN2D | EB11 | NMDAR2D | glutamate receptor, ionotropic, N-methyl D-aspartate 2D | - | HPRD | 10862698 |
| ITSN1 | ITSN | MGC134948 | MGC134949 | SH3D1A | SH3P17 | intersectin 1 (SH3 domain protein) | - | HPRD | 10373452 |
| KIAA1377 | - | KIAA1377 | Two-hybrid | BioGRID | 16169070 |
| MAP3K10 | MLK2 | MST | mitogen-activated protein kinase kinase kinase 10 | Reconstituted Complex | BioGRID | 9742220 |
| MED31 | 3110004H13Rik | CGI-125 | FLJ27436 | FLJ36714 | Soh1 | mediator complex subunit 31 | Two-hybrid | BioGRID | 16169070 |
| MOBKL3 | 2C4D | CGI-95 | MGC12264 | MOB1 | MOB3 | PREI3 | MOB1, Mps One Binder kinase activator-like 3 (yeast) | - | HPRD | 11872741 |
| NCK1 | MGC12668 | NCK | NCKalpha | NCK adaptor protein 1 | - | HPRD,BioGRID | 10206341 |
| NCK2 | GRB4 | NCKbeta | NCK adaptor protein 2 | - | HPRD | 11557983 |
| NME2 | MGC111212 | NDPK-B | NDPKB | NM23-H2 | NM23B | puf | non-metastatic cells 2, protein (NM23B) expressed in | - | HPRD | 11872741 |
| PACSIN1 | KIAA1379 | SDPI | protein kinase C and casein kinase substrate in neurons 1 | - | HPRD,BioGRID | 11082044 |
| PACSIN1 | KIAA1379 | SDPI | protein kinase C and casein kinase substrate in neurons 1 | - | HPRD | 9950691 |11082044 |
| PACSIN2 | SDPII | protein kinase C and casein kinase substrate in neurons 2 | Reconstituted Complex | BioGRID | 11082044 |
| PACSIN3 | SDPIII | protein kinase C and casein kinase substrate in neurons 3 | - | HPRD,BioGRID | 11082044 |
| PFN2 | D3S1319E | PFL | profilin 2 | - | HPRD | 9463375 |
| PIAS1 | DDXBP1 | GBP | GU/RH-II | MGC141878 | MGC141879 | ZMIZ3 | protein inhibitor of activated STAT, 1 | - | HPRD | 15123615 |
| PIN4 | EPVH | MGC138486 | PAR14 | PAR17 | protein (peptidylprolyl cis/trans isomerase) NIMA-interacting, 4 (parvulin) | Two-hybrid | BioGRID | 16169070 |
| PRNP | ASCR | CD230 | CJD | GSS | MGC26679 | PRIP | PrP | PrP27-30 | PrP33-35C | PrPc | prion | prion protein | - | HPRD | 15146195 |
| SH3GL1 | CNSA1 | EEN | MGC111371 | SH3D2B | SH3P8 | SH3-domain GRB2-like 1 | - | HPRD,BioGRID | 9238017 |10764144 |
| SH3GL2 | CNSA2 | EEN-B1 | FLJ20276 | FLJ25015 | SH3D2A | SH3P4 | SH3-domain GRB2-like 2 | Reconstituted Complex | BioGRID | 9341169 |12456676 |
| SH3GL2 | CNSA2 | EEN-B1 | FLJ20276 | FLJ25015 | SH3D2A | SH3P4 | SH3-domain GRB2-like 2 | - | HPRD | 11877424 |
| SH3GL2 | CNSA2 | EEN-B1 | FLJ20276 | FLJ25015 | SH3D2A | SH3P4 | SH3-domain GRB2-like 2 | - | HPRD | 9238017 |11877424 |
| SH3GL3 | CNSA3 | EEN-2B-L3 | EEN-B2 | HsT19371 | SH3D2C | SH3P13 | SH3-domain GRB2-like 3 | Endophilin A3 interacts with Dynamin. | BIND | 11894096 |
| SH3GL3 | CNSA3 | EEN-2B-L3 | EEN-B2 | HsT19371 | SH3D2C | SH3P13 | SH3-domain GRB2-like 3 | - | HPRD | 9238017 |
| SH3GLB1 | Bif-1 | CGI-61 | KIAA0491 | dJ612B15.2 | SH3-domain GRB2-like endophilin B1 | Reconstituted Complex | BioGRID | 12456676 |
| SNAP25 | FLJ23079 | RIC-4 | RIC4 | SEC9 | SNAP | SNAP-25 | bA416N4.2 | dJ1068F16.2 | synaptosomal-associated protein, 25kDa | - | HPRD | 10373452 |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| KEGG ENDOCYTOSIS | 183 | 132 | All SZGR 2.0 genes in this pathway |
| KEGG FC GAMMA R MEDIATED PHAGOCYTOSIS | 97 | 71 | All SZGR 2.0 genes in this pathway |
| BIOCARTA NDKDYNAMIN PATHWAY | 21 | 15 | All SZGR 2.0 genes in this pathway |
| BIOCARTA GABA PATHWAY | 10 | 10 | All SZGR 2.0 genes in this pathway |
| BIOCARTA BARR MAPK PATHWAY | 12 | 11 | All SZGR 2.0 genes in this pathway |
| BIOCARTA BARRESTIN SRC PATHWAY | 15 | 14 | All SZGR 2.0 genes in this pathway |
| BIOCARTA BARRESTIN PATHWAY | 10 | 8 | All SZGR 2.0 genes in this pathway |
| PID NOTCH PATHWAY | 59 | 49 | All SZGR 2.0 genes in this pathway |
| PID EPHB FWD PATHWAY | 40 | 38 | All SZGR 2.0 genes in this pathway |
| PID TXA2PATHWAY | 57 | 43 | All SZGR 2.0 genes in this pathway |
| PID CXCR4 PATHWAY | 102 | 78 | All SZGR 2.0 genes in this pathway |
| PID TRKR PATHWAY | 62 | 48 | All SZGR 2.0 genes in this pathway |
| PID IL8 CXCR2 PATHWAY | 34 | 26 | All SZGR 2.0 genes in this pathway |
| PID ERBB1 INTERNALIZATION PATHWAY | 41 | 35 | All SZGR 2.0 genes in this pathway |
| PID CXCR3 PATHWAY | 43 | 34 | All SZGR 2.0 genes in this pathway |
| PID THROMBIN PAR1 PATHWAY | 43 | 32 | All SZGR 2.0 genes in this pathway |
| PID IL8 CXCR1 PATHWAY | 28 | 19 | All SZGR 2.0 genes in this pathway |
| PID EPHRINB REV PATHWAY | 30 | 25 | All SZGR 2.0 genes in this pathway |
| REACTOME GAP JUNCTION DEGRADATION | 10 | 8 | All SZGR 2.0 genes in this pathway |
| REACTOME SIGNALLING BY NGF | 217 | 167 | All SZGR 2.0 genes in this pathway |
| REACTOME DEVELOPMENTAL BIOLOGY | 396 | 292 | All SZGR 2.0 genes in this pathway |
| REACTOME MEMBRANE TRAFFICKING | 129 | 74 | All SZGR 2.0 genes in this pathway |
| REACTOME NGF SIGNALLING VIA TRKA FROM THE PLASMA MEMBRANE | 137 | 105 | All SZGR 2.0 genes in this pathway |
| REACTOME MHC CLASS II ANTIGEN PRESENTATION | 91 | 61 | All SZGR 2.0 genes in this pathway |
| REACTOME RETROGRADE NEUROTROPHIN SIGNALLING | 13 | 7 | All SZGR 2.0 genes in this pathway |
| REACTOME AXON GUIDANCE | 251 | 188 | All SZGR 2.0 genes in this pathway |
| REACTOME L1CAM INTERACTIONS | 86 | 62 | All SZGR 2.0 genes in this pathway |
| REACTOME RECYCLING PATHWAY OF L1 | 27 | 15 | All SZGR 2.0 genes in this pathway |
| REACTOME INNATE IMMUNE SYSTEM | 279 | 178 | All SZGR 2.0 genes in this pathway |
| REACTOME IMMUNE SYSTEM | 933 | 616 | All SZGR 2.0 genes in this pathway |
| REACTOME TOLL RECEPTOR CASCADES | 118 | 84 | All SZGR 2.0 genes in this pathway |
| REACTOME ADAPTIVE IMMUNE SYSTEM | 539 | 350 | All SZGR 2.0 genes in this pathway |
| REACTOME GAP JUNCTION TRAFFICKING | 27 | 19 | All SZGR 2.0 genes in this pathway |
| CHARAFE BREAST CANCER LUMINAL VS MESENCHYMAL DN | 460 | 312 | All SZGR 2.0 genes in this pathway |
| SENESE HDAC3 TARGETS DN | 536 | 332 | All SZGR 2.0 genes in this pathway |
| ENK UV RESPONSE EPIDERMIS DN | 508 | 354 | All SZGR 2.0 genes in this pathway |
| BERENJENO TRANSFORMED BY RHOA REVERSIBLY UP | 8 | 6 | All SZGR 2.0 genes in this pathway |
| LINDGREN BLADDER CANCER CLUSTER 1 DN | 378 | 231 | All SZGR 2.0 genes in this pathway |
| MARKEY RB1 CHRONIC LOF UP | 115 | 78 | All SZGR 2.0 genes in this pathway |
| GRAESSMANN APOPTOSIS BY SERUM DEPRIVATION DN | 234 | 147 | All SZGR 2.0 genes in this pathway |
| GRAESSMANN RESPONSE TO MC AND SERUM DEPRIVATION DN | 84 | 54 | All SZGR 2.0 genes in this pathway |
| BERENJENO TRANSFORMED BY RHOA DN | 394 | 258 | All SZGR 2.0 genes in this pathway |
| MCBRYAN PUBERTAL BREAST 3 4WK UP | 214 | 144 | All SZGR 2.0 genes in this pathway |
| SHEN SMARCA2 TARGETS DN | 357 | 212 | All SZGR 2.0 genes in this pathway |
| KANG IMMORTALIZED BY TERT DN | 102 | 67 | All SZGR 2.0 genes in this pathway |
| IVANOVA HEMATOPOIESIS STEM CELL LONG TERM | 302 | 191 | All SZGR 2.0 genes in this pathway |
| JIANG AGING HYPOTHALAMUS DN | 40 | 31 | All SZGR 2.0 genes in this pathway |
| DASU IL6 SIGNALING SCAR UP | 30 | 21 | All SZGR 2.0 genes in this pathway |
| BURTON ADIPOGENESIS 7 | 51 | 34 | All SZGR 2.0 genes in this pathway |
| JIANG AGING CEREBRAL CORTEX DN | 54 | 43 | All SZGR 2.0 genes in this pathway |
| BURTON ADIPOGENESIS 9 | 92 | 59 | All SZGR 2.0 genes in this pathway |
| BOQUEST STEM CELL CULTURED VS FRESH UP | 425 | 298 | All SZGR 2.0 genes in this pathway |
| CHANG CORE SERUM RESPONSE DN | 209 | 137 | All SZGR 2.0 genes in this pathway |
| SWEET LUNG CANCER KRAS DN | 435 | 289 | All SZGR 2.0 genes in this pathway |
| VALK AML CLUSTER 11 | 36 | 24 | All SZGR 2.0 genes in this pathway |
| CHIANG LIVER CANCER SUBCLASS INTERFERON DN | 52 | 30 | All SZGR 2.0 genes in this pathway |
| MEISSNER NPC HCP WITH H3K4ME2 | 491 | 319 | All SZGR 2.0 genes in this pathway |
| DASU IL6 SIGNALING UP | 59 | 44 | All SZGR 2.0 genes in this pathway |
| KIM ALL DISORDERS OLIGODENDROCYTE NUMBER CORR UP | 756 | 494 | All SZGR 2.0 genes in this pathway |
| KIM BIPOLAR DISORDER OLIGODENDROCYTE DENSITY CORR UP | 682 | 440 | All SZGR 2.0 genes in this pathway |
| KIM ALL DISORDERS CALB1 CORR UP | 548 | 370 | All SZGR 2.0 genes in this pathway |
| CHICAS RB1 TARGETS SENESCENT | 572 | 352 | All SZGR 2.0 genes in this pathway |
| JOHNSTONE PARVB TARGETS 3 UP | 430 | 288 | All SZGR 2.0 genes in this pathway |
| BILANGES SERUM SENSITIVE GENES | 90 | 54 | All SZGR 2.0 genes in this pathway |
| BILANGES SERUM RESPONSE TRANSLATION | 37 | 20 | All SZGR 2.0 genes in this pathway |
| SERVITJA ISLET HNF1A TARGETS UP | 163 | 111 | All SZGR 2.0 genes in this pathway |
| DELACROIX RARG BOUND MEF | 367 | 231 | All SZGR 2.0 genes in this pathway |
| PECE MAMMARY STEM CELL DN | 146 | 88 | All SZGR 2.0 genes in this pathway |
Section VI. microRNA annotation
| miRNA family | Target position | miRNA ID | miRNA seq | ||
|---|---|---|---|---|---|
| UTR start | UTR end | Match method | |||
| miR-224 | 107 | 114 | 1A,m8 | hsa-miR-224 | CAAGUCACUAGUGGUUCCGUUUA |
| hsa-miR-224 | CAAGUCACUAGUGGUUCCGUUUA | ||||
- SZ: miRNAs which differentially expressed in brain cortex of schizophrenia patients comparing with control samples using microarray. Click here to see the list of SZ related miRNAs.
- Brain: miRNAs which are expressed in brain based on miRNA microarray expression studies. Click here to see the list of brain related miRNAs.