Gene Page: ABCA1
Summary ?
| GeneID | 19 |
| Symbol | ABCA1 |
| Synonyms | ABC-1|ABC1|CERP|HDLDT1|TGD |
| Description | ATP binding cassette subfamily A member 1 |
| Reference | MIM:600046|HGNC:HGNC:29|Ensembl:ENSG00000165029|HPRD:02501|Vega:OTTHUMG00000020417 |
| Gene type | protein-coding |
| Map location | 9q31.1 |
| Pascal p-value | 0.243 |
| Sherlock p-value | 0.383 |
| DEG p-value | DEG:Maycox_2009:CC_BA10_fold_change=1.14:CC_BA10_disease_P=0.0337:HBB_BA9_fold_change=1.28:HBB_BA9_disease_P=0.0220 |
| Fetal beta | 1.893 |
| DMG | 1 (# studies) |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CV:GWASdb | Genome-wide Association Studies | GWASdb records for schizophrenia | |
| CV:PGCnp | Genome-wide Association Study | GWAS | |
| DEG:Maycox_2009 | Microarray to determine the expression of over 30000 mRNA transcripts in post-mortem tissue | We included 51 genes whose expression changes are common between two schizophrenia cohorts. | |
| DMG:vanEijk_2014 | Genome-wide DNA methylation analysis | This dataset includes 432 differentially methylated CpG sites corresponding to 391 unique transcripts between schizophrenia patients (n=260) and unaffected controls (n=250). | 1 |
| PMID:cooccur | High-throughput literature-search | Systematic search in PubMed for genes co-occurring with SCZ keywords. A total of 3027 genes were included. | |
| Literature | High-throughput literature-search | Co-occurance with Schizophrenia keywords: schizophrenia,schizophrenias | Click to show details |
| GO_Annotation | Mapping neuro-related keywords to Gene Ontology annotations | Hits with neuro-related keywords: 1 | |
| Network | Shortest path distance of core genes in the Human protein-protein interaction network | Contribution to shortest path in PPI network: 0.0323 |
Section I. Genetics and epigenetics annotation
 Differentially methylated gene
Differentially methylated gene
| Probe | Chromosome | Position | Nearest gene | P (dis) | Beta (dis) | FDR (dis) | Study |
|---|---|---|---|---|---|---|---|
| cg13160888 | 9 | 107526516 | ABCA1 | 0.016 | -7.686 | DMG:vanEijk_2014 |
Section II. Transcriptome annotation
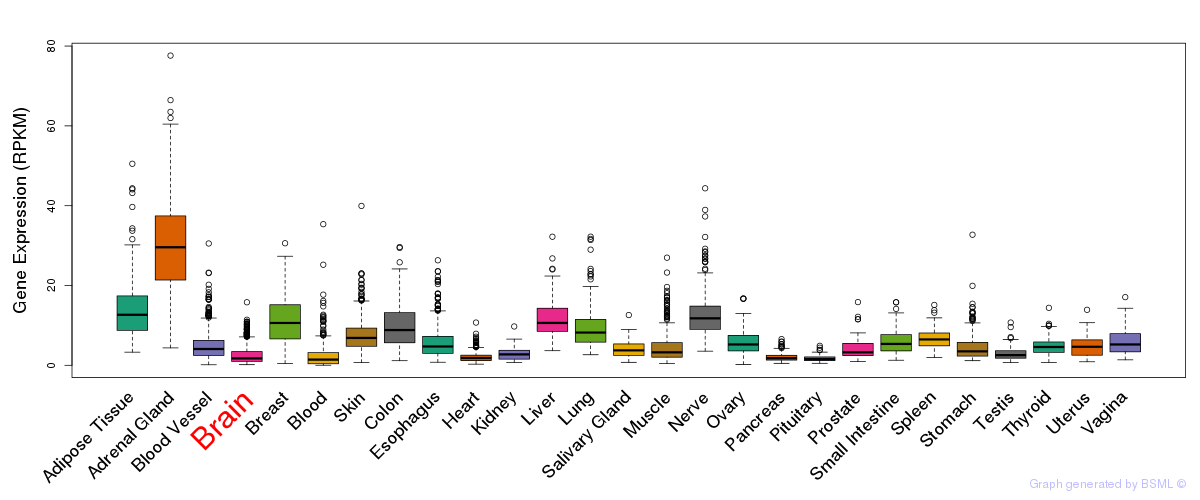
General gene expression (GTEx)
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
Top co-expressed genes in brain regions
| Top 10 positively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| DHX15 | 0.94 | 0.91 |
| SF3B1 | 0.93 | 0.92 |
| ZNF22 | 0.93 | 0.90 |
| ELP2 | 0.93 | 0.91 |
| CDKAL1 | 0.93 | 0.89 |
| METAP2 | 0.92 | 0.92 |
| C2orf67 | 0.92 | 0.91 |
| DHX9 | 0.92 | 0.88 |
| DDX5 | 0.92 | 0.91 |
| UHRF2 | 0.92 | 0.89 |
| Top 10 negatively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| AF347015.27 | -0.70 | -0.81 |
| MT-CO2 | -0.69 | -0.82 |
| AF347015.31 | -0.69 | -0.82 |
| IFI27 | -0.67 | -0.82 |
| AF347015.33 | -0.67 | -0.78 |
| HLA-F | -0.67 | -0.72 |
| AF347015.8 | -0.67 | -0.81 |
| MT-CYB | -0.67 | -0.80 |
| AIFM3 | -0.66 | -0.72 |
| PTH1R | -0.66 | -0.73 |
Section III. Gene Ontology annotation
| Molecular function | GO term | Evidence | Neuro keywords | PubMed ID |
|---|---|---|---|---|
| GO:0000166 | nucleotide binding | IEA | - | |
| GO:0005543 | phospholipid binding | IC | 16702602 | |
| GO:0005548 | phospholipid transporter activity | IDA | 16702602 | |
| GO:0005515 | protein binding | IPI | 16192269 | |
| GO:0005524 | ATP binding | IEA | - | |
| GO:0008509 | anion transmembrane transporter activity | IEA | - | |
| GO:0017127 | cholesterol transporter activity | IDA | 12084722 | |
| GO:0016887 | ATPase activity | IEA | - | |
| GO:0015485 | cholesterol binding | IC | 12084722 | |
| GO:0031267 | small GTPase binding | IPI | 16443932 | |
| GO:0030349 | syntaxin-13 binding | IPI | 15469992 | |
| GO:0034188 | apolipoprotein A-I receptor activity | IDA | 16443932 | |
| Biological process | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0060155 | platelet dense granule organization | IMP | serotonin (GO term level: 7) | 15163665 |
| GO:0002790 | peptide secretion | IEA | - | |
| GO:0007186 | G-protein coupled receptor protein signaling pathway | IMP | 16443932 | |
| GO:0006497 | protein amino acid lipidation | IEA | - | |
| GO:0008202 | steroid metabolic process | IEA | - | |
| GO:0008203 | cholesterol metabolic process | IEA | - | |
| GO:0006810 | transport | IEA | - | |
| GO:0007040 | lysosome organization | IDA | 15163665 | |
| GO:0006629 | lipid metabolic process | IEA | - | |
| GO:0006911 | phagocytosis, engulfment | IEA | - | |
| GO:0016197 | endosome transport | IDA | 14747463 | |
| GO:0033344 | cholesterol efflux | IEA | - | |
| GO:0032367 | intracellular cholesterol transport | IMP | 10431236 | |
| GO:0030819 | positive regulation of cAMP biosynthetic process | IMP | 14701824 | |
| GO:0032488 | Cdc42 protein signal transduction | IMP | 16443932 | |
| GO:0033700 | phospholipid efflux | IDA | 10431236 |11162594 | |
| GO:0033700 | phospholipid efflux | IMP | 16702602 | |
| GO:0042632 | cholesterol homeostasis | IDA | 10431236 | |
| GO:0045332 | phospholipid translocation | IEA | - | |
| GO:0050702 | interleukin-1 beta secretion | IMP | 11855831 | |
| GO:0043691 | reverse cholesterol transport | IEA | - | |
| GO:0055091 | phospholipid homeostasis | IMP | 16702602 | |
| Cellular component | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0005794 | Golgi apparatus | IEA | - | |
| GO:0005624 | membrane fraction | IDA | 10525055 | |
| GO:0005886 | plasma membrane | IEA | - | |
| GO:0005887 | integral to plasma membrane | IEA | - | |
| GO:0045335 | phagocytic vesicle | IDA | 15469992 | |
| GO:0045121 | membrane raft | IDA | 15469992 |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| KEGG ABC TRANSPORTERS | 44 | 29 | All SZGR 2.0 genes in this pathway |
| PID RXR VDR PATHWAY | 26 | 24 | All SZGR 2.0 genes in this pathway |
| REACTOME PPARA ACTIVATES GENE EXPRESSION | 104 | 72 | All SZGR 2.0 genes in this pathway |
| REACTOME HDL MEDIATED LIPID TRANSPORT | 15 | 12 | All SZGR 2.0 genes in this pathway |
| REACTOME METABOLISM OF LIPIDS AND LIPOPROTEINS | 478 | 302 | All SZGR 2.0 genes in this pathway |
| REACTOME FATTY ACID TRIACYLGLYCEROL AND KETONE BODY METABOLISM | 168 | 115 | All SZGR 2.0 genes in this pathway |
| REACTOME LIPID DIGESTION MOBILIZATION AND TRANSPORT | 46 | 37 | All SZGR 2.0 genes in this pathway |
| REACTOME LIPOPROTEIN METABOLISM | 28 | 24 | All SZGR 2.0 genes in this pathway |
| NAKAMURA TUMOR ZONE PERIPHERAL VS CENTRAL UP | 285 | 181 | All SZGR 2.0 genes in this pathway |
| ZHOU INFLAMMATORY RESPONSE LIVE UP | 485 | 293 | All SZGR 2.0 genes in this pathway |
| SAMOLS TARGETS OF KHSV MIRNAS DN | 62 | 35 | All SZGR 2.0 genes in this pathway |
| CASORELLI ACUTE PROMYELOCYTIC LEUKEMIA UP | 177 | 110 | All SZGR 2.0 genes in this pathway |
| HUTTMANN B CLL POOR SURVIVAL DN | 60 | 39 | All SZGR 2.0 genes in this pathway |
| HORIUCHI WTAP TARGETS UP | 306 | 188 | All SZGR 2.0 genes in this pathway |
| SMIRNOV CIRCULATING ENDOTHELIOCYTES IN CANCER UP | 158 | 103 | All SZGR 2.0 genes in this pathway |
| HOEBEKE LYMPHOID STEM CELL UP | 95 | 64 | All SZGR 2.0 genes in this pathway |
| RHEIN ALL GLUCOCORTICOID THERAPY UP | 78 | 41 | All SZGR 2.0 genes in this pathway |
| KINSEY TARGETS OF EWSR1 FLII FUSION DN | 329 | 219 | All SZGR 2.0 genes in this pathway |
| MARKEY RB1 ACUTE LOF UP | 215 | 137 | All SZGR 2.0 genes in this pathway |
| GAUSSMANN MLL AF4 FUSION TARGETS E UP | 97 | 60 | All SZGR 2.0 genes in this pathway |
| PEREZ TP53 TARGETS | 1174 | 695 | All SZGR 2.0 genes in this pathway |
| HERNANDEZ MITOTIC ARREST BY DOCETAXEL 2 UP | 64 | 45 | All SZGR 2.0 genes in this pathway |
| XU HGF TARGETS REPRESSED BY AKT1 DN | 95 | 58 | All SZGR 2.0 genes in this pathway |
| DAUER STAT3 TARGETS UP | 49 | 35 | All SZGR 2.0 genes in this pathway |
| NUYTTEN NIPP1 TARGETS DN | 848 | 527 | All SZGR 2.0 genes in this pathway |
| BUYTAERT PHOTODYNAMIC THERAPY STRESS UP | 811 | 508 | All SZGR 2.0 genes in this pathway |
| PASQUALUCCI LYMPHOMA BY GC STAGE UP | 283 | 177 | All SZGR 2.0 genes in this pathway |
| MORI MATURE B LYMPHOCYTE UP | 90 | 62 | All SZGR 2.0 genes in this pathway |
| BYSTRYKH HEMATOPOIESIS STEM CELL AND BRAIN QTL TRANS | 185 | 114 | All SZGR 2.0 genes in this pathway |
| MANALO HYPOXIA UP | 207 | 145 | All SZGR 2.0 genes in this pathway |
| YU MYC TARGETS DN | 55 | 38 | All SZGR 2.0 genes in this pathway |
| GERY CEBP TARGETS | 126 | 90 | All SZGR 2.0 genes in this pathway |
| AFFAR YY1 TARGETS UP | 214 | 133 | All SZGR 2.0 genes in this pathway |
| BLALOCK ALZHEIMERS DISEASE INCIPIENT UP | 390 | 242 | All SZGR 2.0 genes in this pathway |
| RUAN RESPONSE TO TNF UP | 12 | 10 | All SZGR 2.0 genes in this pathway |
| BLALOCK ALZHEIMERS DISEASE UP | 1691 | 1088 | All SZGR 2.0 genes in this pathway |
| BURTON ADIPOGENESIS 3 | 101 | 64 | All SZGR 2.0 genes in this pathway |
| RUAN RESPONSE TO TNF TROGLITAZONE UP | 17 | 11 | All SZGR 2.0 genes in this pathway |
| MCLACHLAN DENTAL CARIES DN | 245 | 144 | All SZGR 2.0 genes in this pathway |
| SARTIPY NORMAL AT INSULIN RESISTANCE DN | 21 | 11 | All SZGR 2.0 genes in this pathway |
| WANG SMARCE1 TARGETS UP | 280 | 183 | All SZGR 2.0 genes in this pathway |
| HAN JNK SINGALING DN | 39 | 27 | All SZGR 2.0 genes in this pathway |
| MCLACHLAN DENTAL CARIES UP | 253 | 147 | All SZGR 2.0 genes in this pathway |
| BURTON ADIPOGENESIS 6 | 189 | 112 | All SZGR 2.0 genes in this pathway |
| BILD HRAS ONCOGENIC SIGNATURE | 261 | 166 | All SZGR 2.0 genes in this pathway |
| JEPSEN SMRT TARGETS | 33 | 23 | All SZGR 2.0 genes in this pathway |
| MASSARWEH TAMOXIFEN RESISTANCE UP | 578 | 341 | All SZGR 2.0 genes in this pathway |
| BOCHKIS FOXA2 TARGETS | 425 | 261 | All SZGR 2.0 genes in this pathway |
| MATZUK POST-IMPLANTATION AND POST-PARTUM | 14 | 13 | All SZGR 2.0 genes in this pathway |
| WEST ADRENOCORTICAL TUMOR DN | 546 | 362 | All SZGR 2.0 genes in this pathway |
| BREDEMEYER RAG SIGNALING NOT VIA ATM DN | 57 | 34 | All SZGR 2.0 genes in this pathway |
| BEIER GLIOMA STEM CELL UP | 39 | 17 | All SZGR 2.0 genes in this pathway |
| SWEET LUNG CANCER KRAS DN | 435 | 289 | All SZGR 2.0 genes in this pathway |
| HOFFMANN LARGE TO SMALL PRE BII LYMPHOCYTE DN | 72 | 47 | All SZGR 2.0 genes in this pathway |
| MCMURRAY TP53 HRAS COOPERATION RESPONSE DN | 67 | 46 | All SZGR 2.0 genes in this pathway |
| WONG ADULT TISSUE STEM MODULE | 721 | 492 | All SZGR 2.0 genes in this pathway |
| YAO TEMPORAL RESPONSE TO PROGESTERONE CLUSTER 0 | 76 | 54 | All SZGR 2.0 genes in this pathway |
| NAKAMURA ADIPOGENESIS EARLY UP | 66 | 44 | All SZGR 2.0 genes in this pathway |
| NAKAMURA ADIPOGENESIS LATE UP | 104 | 67 | All SZGR 2.0 genes in this pathway |
| HIRSCH CELLULAR TRANSFORMATION SIGNATURE UP | 242 | 159 | All SZGR 2.0 genes in this pathway |
| DUTERTRE ESTRADIOL RESPONSE 24HR DN | 505 | 328 | All SZGR 2.0 genes in this pathway |
| WIERENGA STAT5A TARGETS UP | 217 | 131 | All SZGR 2.0 genes in this pathway |
| WIERENGA STAT5A TARGETS GROUP2 | 60 | 38 | All SZGR 2.0 genes in this pathway |
| PILON KLF1 TARGETS UP | 504 | 321 | All SZGR 2.0 genes in this pathway |
| JOHNSTONE PARVB TARGETS 3 UP | 430 | 288 | All SZGR 2.0 genes in this pathway |
| LEE BMP2 TARGETS UP | 745 | 475 | All SZGR 2.0 genes in this pathway |
| PURBEY TARGETS OF CTBP1 NOT SATB1 UP | 344 | 215 | All SZGR 2.0 genes in this pathway |
| GOBERT CORE OLIGODENDROCYTE DIFFERENTIATION | 40 | 28 | All SZGR 2.0 genes in this pathway |
| SCHMIDT POR TARGETS IN LIMB BUD DN | 8 | 7 | All SZGR 2.0 genes in this pathway |
| YANG BCL3 TARGETS UP | 364 | 236 | All SZGR 2.0 genes in this pathway |
| PHONG TNF RESPONSE NOT VIA P38 | 337 | 236 | All SZGR 2.0 genes in this pathway |
| RAO BOUND BY SALL4 ISOFORM A | 182 | 108 | All SZGR 2.0 genes in this pathway |
Section VI. microRNA annotation
| miRNA family | Target position | miRNA ID | miRNA seq | ||
|---|---|---|---|---|---|
| UTR start | UTR end | Match method | |||
| miR-101 | 2687 | 2693 | 1A | hsa-miR-101 | UACAGUACUGUGAUAACUGAAG |
| miR-124/506 | 185 | 191 | m8 | hsa-miR-506 | UAAGGCACCCUUCUGAGUAGA |
| hsa-miR-124brain | UAAGGCACGCGGUGAAUGCC | ||||
| miR-128 | 2593 | 2599 | 1A | hsa-miR-128a | UCACAGUGAACCGGUCUCUUUU |
| hsa-miR-128b | UCACAGUGAACCGGUCUCUUUC | ||||
| miR-130/301 | 3111 | 3117 | m8 | hsa-miR-130abrain | CAGUGCAAUGUUAAAAGGGCAU |
| hsa-miR-301 | CAGUGCAAUAGUAUUGUCAAAGC | ||||
| hsa-miR-130bbrain | CAGUGCAAUGAUGAAAGGGCAU | ||||
| hsa-miR-454-3p | UAGUGCAAUAUUGCUUAUAGGGUUU | ||||
| miR-142-5p | 3223 | 3230 | 1A,m8 | hsa-miR-142-5p | CAUAAAGUAGAAAGCACUAC |
| miR-144 | 2687 | 2693 | 1A | hsa-miR-144 | UACAGUAUAGAUGAUGUACUAG |
| miR-145 | 3115 | 3121 | 1A | hsa-miR-145 | GUCCAGUUUUCCCAGGAAUCCCUU |
| hsa-miR-145 | GUCCAGUUUUCCCAGGAAUCCCUU | ||||
| miR-148/152 | 3112 | 3118 | m8 | hsa-miR-148a | UCAGUGCACUACAGAACUUUGU |
| hsa-miR-152brain | UCAGUGCAUGACAGAACUUGGG | ||||
| hsa-miR-148b | UCAGUGCAUCACAGAACUUUGU | ||||
| miR-17-5p/20/93.mr/106/519.d | 3079 | 3086 | 1A,m8 | hsa-miR-17-5p | CAAAGUGCUUACAGUGCAGGUAGU |
| hsa-miR-20abrain | UAAAGUGCUUAUAGUGCAGGUAG | ||||
| hsa-miR-106a | AAAAGUGCUUACAGUGCAGGUAGC | ||||
| hsa-miR-106bSZ | UAAAGUGCUGACAGUGCAGAU | ||||
| hsa-miR-20bSZ | CAAAGUGCUCAUAGUGCAGGUAG | ||||
| hsa-miR-519d | CAAAGUGCCUCCCUUUAGAGUGU | ||||
| miR-183 | 1367 | 1373 | m8 | hsa-miR-183 | UAUGGCACUGGUAGAAUUCACUG |
| miR-19 | 3110 | 3116 | m8 | hsa-miR-19a | UGUGCAAAUCUAUGCAAAACUGA |
| hsa-miR-19b | UGUGCAAAUCCAUGCAAAACUGA | ||||
| miR-199 | 3114 | 3120 | 1A | hsa-miR-199a | CCCAGUGUUCAGACUACCUGUUC |
| hsa-miR-199b | CCCAGUGUUUAGACUAUCUGUUC | ||||
| miR-23 | 255 | 261 | 1A | hsa-miR-23abrain | AUCACAUUGCCAGGGAUUUCC |
| hsa-miR-23bbrain | AUCACAUUGCCAGGGAUUACC | ||||
| miR-26 | 227 | 233 | 1A | hsa-miR-26abrain | UUCAAGUAAUCCAGGAUAGGC |
| hsa-miR-26bSZ | UUCAAGUAAUUCAGGAUAGGUU | ||||
| miR-27 | 2593 | 2600 | 1A,m8 | hsa-miR-27abrain | UUCACAGUGGCUAAGUUCCGC |
| hsa-miR-27bbrain | UUCACAGUGGCUAAGUUCUGC | ||||
| miR-324-3p | 181 | 187 | m8 | hsa-miR-324-3p | CCACUGCCCCAGGUGCUGCUGG |
| miR-33 | 150 | 157 | 1A,m8 | hsa-miR-33 | GUGCAUUGUAGUUGCAUUG |
| hsa-miR-33b | GUGCAUUGCUGUUGCAUUGCA | ||||
| hsa-miR-33 | GUGCAUUGUAGUUGCAUUG | ||||
| hsa-miR-33b | GUGCAUUGCUGUUGCAUUGCA | ||||
| hsa-miR-33 | GUGCAUUGUAGUUGCAUUG | ||||
| hsa-miR-33b | GUGCAUUGCUGUUGCAUUGCA | ||||
| miR-376c | 3252 | 3258 | m8 | hsa-miR-376c | AACAUAGAGGAAAUUCCACG |
| miR-381 | 2808 | 2814 | 1A | hsa-miR-381 | UAUACAAGGGCAAGCUCUCUGU |
| hsa-miR-381 | UAUACAAGGGCAAGCUCUCUGU | ||||
| miR-9 | 2833 | 2839 | 1A | hsa-miR-9SZ | UCUUUGGUUAUCUAGCUGUAUGA |
| miR-93.hd/291-3p/294/295/302/372/373/520 | 3078 | 3084 | m8 | hsa-miR-93brain | AAAGUGCUGUUCGUGCAGGUAG |
| hsa-miR-302a | UAAGUGCUUCCAUGUUUUGGUGA | ||||
| hsa-miR-302b | UAAGUGCUUCCAUGUUUUAGUAG | ||||
| hsa-miR-302c | UAAGUGCUUCCAUGUUUCAGUGG | ||||
| hsa-miR-302d | UAAGUGCUUCCAUGUUUGAGUGU | ||||
| hsa-miR-372 | AAAGUGCUGCGACAUUUGAGCGU | ||||
| hsa-miR-373 | GAAGUGCUUCGAUUUUGGGGUGU | ||||
| hsa-miR-520e | AAAGUGCUUCCUUUUUGAGGG | ||||
| hsa-miR-520a | AAAGUGCUUCCCUUUGGACUGU | ||||
| hsa-miR-520b | AAAGUGCUUCCUUUUAGAGGG | ||||
| hsa-miR-520c | AAAGUGCUUCCUUUUAGAGGGUU | ||||
| hsa-miR-520d | AAAGUGCUUCUCUUUGGUGGGUU | ||||
| miR-96 | 499 | 505 | m8 | hsa-miR-96brain | UUUGGCACUAGCACAUUUUUGC |
- SZ: miRNAs which differentially expressed in brain cortex of schizophrenia patients comparing with control samples using microarray. Click here to see the list of SZ related miRNAs.
- Brain: miRNAs which are expressed in brain based on miRNA microarray expression studies. Click here to see the list of brain related miRNAs.