| KEGG NUCLEOTIDE EXCISION REPAIR
| 44 | 25 | All SZGR 2.0 genes in this pathway |
| REACTOME RNA POL I TRANSCRIPTION TERMINATION
| 22 | 12 | All SZGR 2.0 genes in this pathway |
| REACTOME RNA POL I TRANSCRIPTION
| 89 | 64 | All SZGR 2.0 genes in this pathway |
| REACTOME RNA POL II TRANSCRIPTION
| 105 | 54 | All SZGR 2.0 genes in this pathway |
| REACTOME MRNA CAPPING
| 30 | 11 | All SZGR 2.0 genes in this pathway |
| REACTOME TRANSCRIPTION COUPLED NER TC NER
| 45 | 24 | All SZGR 2.0 genes in this pathway |
| REACTOME RNA POL II TRANSCRIPTION PRE INITIATION AND PROMOTER OPENING
| 41 | 21 | All SZGR 2.0 genes in this pathway |
| REACTOME MRNA PROCESSING
| 161 | 86 | All SZGR 2.0 genes in this pathway |
| REACTOME TRANSCRIPTION
| 210 | 127 | All SZGR 2.0 genes in this pathway |
| REACTOME NUCLEOTIDE EXCISION REPAIR
| 51 | 27 | All SZGR 2.0 genes in this pathway |
| REACTOME FORMATION OF RNA POL II ELONGATION COMPLEX
| 45 | 19 | All SZGR 2.0 genes in this pathway |
| REACTOME FORMATION OF TRANSCRIPTION COUPLED NER TC NER REPAIR COMPLEX
| 30 | 13 | All SZGR 2.0 genes in this pathway |
| REACTOME RNA POL I RNA POL III AND MITOCHONDRIAL TRANSCRIPTION
| 122 | 80 | All SZGR 2.0 genes in this pathway |
| REACTOME DNA REPAIR
| 112 | 59 | All SZGR 2.0 genes in this pathway |
| REACTOME RNA POL II PRE TRANSCRIPTION EVENTS
| 61 | 32 | All SZGR 2.0 genes in this pathway |
| REACTOME GLOBAL GENOMIC NER GG NER
| 35 | 20 | All SZGR 2.0 genes in this pathway |
| REACTOME FORMATION OF INCISION COMPLEX IN GG NER
| 23 | 11 | All SZGR 2.0 genes in this pathway |
| REACTOME HIV INFECTION
| 207 | 122 | All SZGR 2.0 genes in this pathway |
| REACTOME HIV LIFE CYCLE
| 125 | 69 | All SZGR 2.0 genes in this pathway |
| REACTOME FORMATION OF THE HIV1 EARLY ELONGATION COMPLEX
| 34 | 11 | All SZGR 2.0 genes in this pathway |
| REACTOME LATE PHASE OF HIV LIFE CYCLE
| 104 | 61 | All SZGR 2.0 genes in this pathway |
| REACTOME RNA POL I TRANSCRIPTION INITIATION
| 25 | 14 | All SZGR 2.0 genes in this pathway |
| DACOSTA UV RESPONSE VIA ERCC3 TTD UP
| 64 | 39 | All SZGR 2.0 genes in this pathway |
| ROVERSI GLIOMA COPY NUMBER UP
| 100 | 75 | All SZGR 2.0 genes in this pathway |
| SCHLOSSER SERUM RESPONSE UP
| 134 | 93 | All SZGR 2.0 genes in this pathway |
| DACOSTA UV RESPONSE VIA ERCC3 UP
| 309 | 199 | All SZGR 2.0 genes in this pathway |
| PUJANA ATM PCC NETWORK
| 1442 | 892 | All SZGR 2.0 genes in this pathway |
| KOYAMA SEMA3B TARGETS UP
| 292 | 168 | All SZGR 2.0 genes in this pathway |
| KAUFFMANN DNA REPAIR GENES
| 230 | 137 | All SZGR 2.0 genes in this pathway |
| BYSTROEM CORRELATED WITH IL5 DN
| 64 | 47 | All SZGR 2.0 genes in this pathway |
| XU CREBBP TARGETS DN
| 44 | 31 | All SZGR 2.0 genes in this pathway |
| SU PANCREAS
| 54 | 30 | All SZGR 2.0 genes in this pathway |
| JOSEPH RESPONSE TO SODIUM BUTYRATE DN
| 64 | 45 | All SZGR 2.0 genes in this pathway |
| BROWNE HCMV INFECTION 24HR DN
| 148 | 102 | All SZGR 2.0 genes in this pathway |
| BROWNE HCMV INFECTION 14HR DN
| 298 | 200 | All SZGR 2.0 genes in this pathway |
| DAZARD RESPONSE TO UV NHEK UP
| 244 | 151 | All SZGR 2.0 genes in this pathway |
| ZHANG RESPONSE TO CANTHARIDIN DN
| 69 | 46 | All SZGR 2.0 genes in this pathway |
| DAZARD UV RESPONSE CLUSTER G1
| 67 | 41 | All SZGR 2.0 genes in this pathway |
| BROWNE HCMV INFECTION 12HR DN
| 101 | 64 | All SZGR 2.0 genes in this pathway |
| IWANAGA CARCINOGENESIS BY KRAS PTEN UP
| 181 | 112 | All SZGR 2.0 genes in this pathway |
| DAIRKEE CANCER PRONE RESPONSE E2
| 28 | 21 | All SZGR 2.0 genes in this pathway |
| MATZUK MEIOTIC AND DNA REPAIR
| 39 | 26 | All SZGR 2.0 genes in this pathway |
| GRESHOCK CANCER COPY NUMBER UP
| 323 | 240 | All SZGR 2.0 genes in this pathway |
| YAO TEMPORAL RESPONSE TO PROGESTERONE CLUSTER 4
| 15 | 11 | All SZGR 2.0 genes in this pathway |
| VERHAAK GLIOBLASTOMA CLASSICAL
| 162 | 122 | All SZGR 2.0 genes in this pathway |
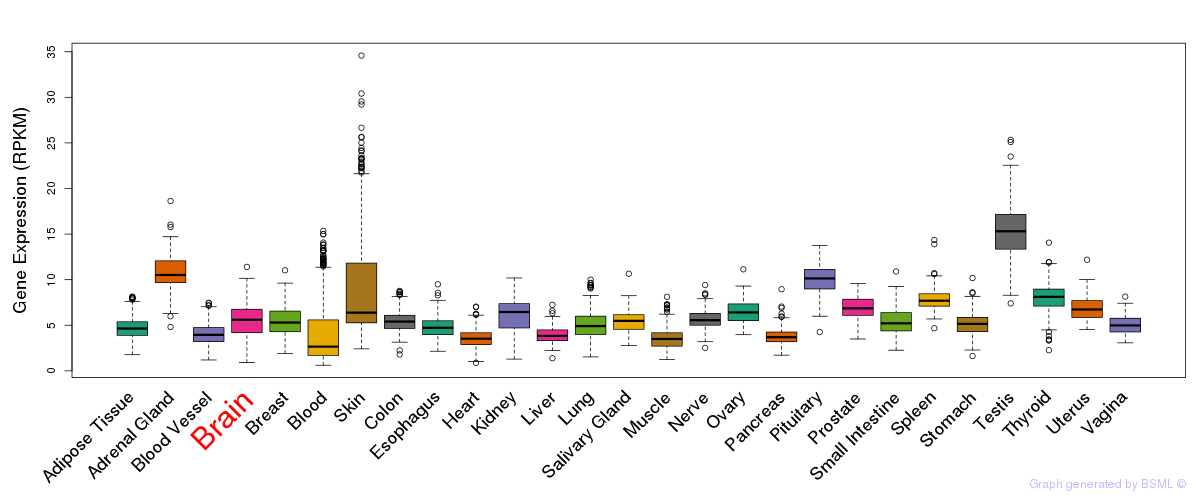
 Differentially methylated gene
Differentially methylated gene eQTL annotation
eQTL annotation