Gene Page: FGD2
Summary ?
| GeneID | 221472 |
| Symbol | FGD2 |
| Synonyms | ZFYVE4 |
| Description | FYVE, RhoGEF and PH domain containing 2 |
| Reference | MIM:605091|HGNC:HGNC:3664|Ensembl:ENSG00000146192|HPRD:05478|Vega:OTTHUMG00000014616 |
| Gene type | protein-coding |
| Map location | 6p21.2 |
| Pascal p-value | 0.358 |
| DEG p-value | DEG:Sanders_2014:DS1_p=-0.151:DS1_beta=0.033300:DS2_p=4.97e-01:DS2_beta=-0.034:DS2_FDR=7.25e-01 |
| eGene | Myers' cis & trans Meta |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CV:GWASdb | Genome-wide Association Studies | GWASdb records for schizophrenia | |
| CV:PGCnp | Genome-wide Association Study | GWAS | |
| DEG:Sanders_2013 | Microarray | Whole-genome gene expression profiles using microarrays on lymphoblastoid cell lines (LCLs) from 413 cases and 446 controls. | |
| GSMA_I | Genome scan meta-analysis | Psr: 0.033 | |
| GSMA_IIE | Genome scan meta-analysis (European-ancestry samples) | Psr: 0.04433 | |
| GO_Annotation | Mapping neuro-related keywords to Gene Ontology annotations | Hits with neuro-related keywords: 1 |
Section I. Genetics and epigenetics annotation
 eQTL annotation
eQTL annotation
| SNP ID | Chromosome | Position | eGene | Gene Entrez ID | pvalue | qvalue | TSS distance | eQTL type |
|---|---|---|---|---|---|---|---|---|
| rs9366917 | chr6 | 36790665 | FGD2 | 221472 | 0.04 | cis |
Section II. Transcriptome annotation
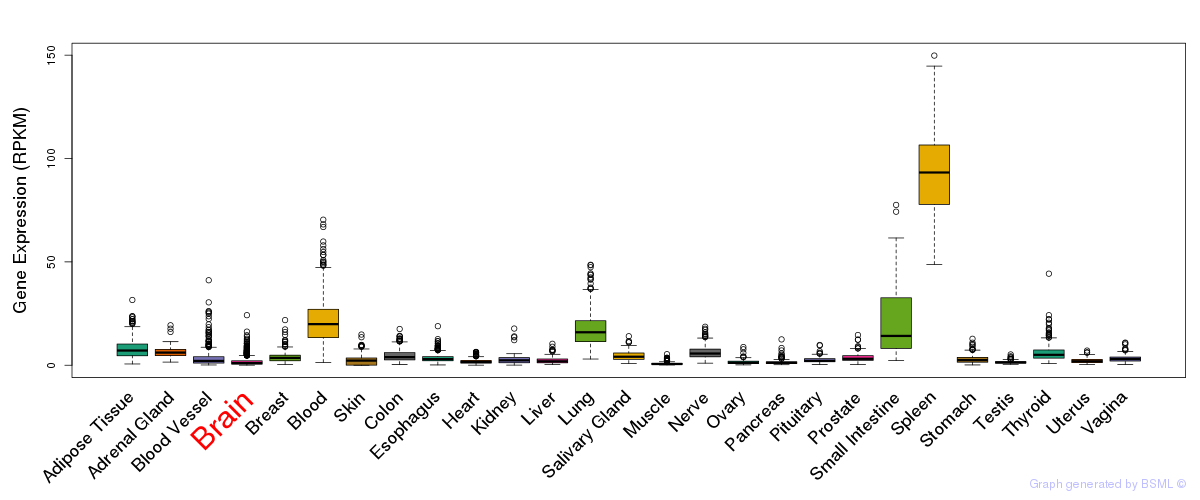
General gene expression (GTEx)
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
No co-expressed genes in brain regions
Section III. Gene Ontology annotation
| Molecular function | GO term | Evidence | Neuro keywords | PubMed ID |
|---|---|---|---|---|
| GO:0005089 | Rho guanyl-nucleotide exchange factor activity | IEA | - | |
| GO:0005085 | guanyl-nucleotide exchange factor activity | ISS | - | |
| GO:0005515 | protein binding | ISS | - | |
| GO:0008270 | zinc ion binding | IEA | - | |
| GO:0031267 | small GTPase binding | ISS | - | |
| GO:0046872 | metal ion binding | IEA | - | |
| Biological process | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0046847 | filopodium formation | ISS | axon (GO term level: 10) | - |
| GO:0007010 | cytoskeleton organization | ISS | - | |
| GO:0008360 | regulation of cell shape | ISS | - | |
| GO:0030036 | actin cytoskeleton organization | ISS | - | |
| GO:0035023 | regulation of Rho protein signal transduction | IEA | - | |
| GO:0043088 | regulation of Cdc42 GTPase activity | ISS | - | |
| Cellular component | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0001726 | ruffle | ISS | - | |
| GO:0005794 | Golgi apparatus | ISS | - | |
| GO:0005856 | cytoskeleton | IEA | - | |
| GO:0005622 | intracellular | IEA | - | |
| GO:0005737 | cytoplasm | ISS | - | |
| GO:0030027 | lamellipodium | ISS | - |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| REACTOME SIGNALING BY RHO GTPASES | 113 | 81 | All SZGR 2.0 genes in this pathway |
| REACTOME SIGNALLING BY NGF | 217 | 167 | All SZGR 2.0 genes in this pathway |
| REACTOME NRAGE SIGNALS DEATH THROUGH JNK | 43 | 33 | All SZGR 2.0 genes in this pathway |
| REACTOME CELL DEATH SIGNALLING VIA NRAGE NRIF AND NADE | 60 | 43 | All SZGR 2.0 genes in this pathway |
| REACTOME P75 NTR RECEPTOR MEDIATED SIGNALLING | 81 | 61 | All SZGR 2.0 genes in this pathway |
| REACTOME SIGNALING BY GPCR | 920 | 449 | All SZGR 2.0 genes in this pathway |
| REACTOME G ALPHA1213 SIGNALLING EVENTS | 74 | 56 | All SZGR 2.0 genes in this pathway |
| REACTOME GPCR DOWNSTREAM SIGNALING | 805 | 368 | All SZGR 2.0 genes in this pathway |
| DODD NASOPHARYNGEAL CARCINOMA UP | 1821 | 933 | All SZGR 2.0 genes in this pathway |
| COATES MACROPHAGE M1 VS M2 UP | 81 | 52 | All SZGR 2.0 genes in this pathway |
| LABBE TARGETS OF TGFB1 AND WNT3A DN | 108 | 68 | All SZGR 2.0 genes in this pathway |
| SENGUPTA EBNA1 ANTICORRELATED | 173 | 85 | All SZGR 2.0 genes in this pathway |
| WANG RESPONSE TO GSK3 INHIBITOR SB216763 UP | 397 | 206 | All SZGR 2.0 genes in this pathway |
| KASLER HDAC7 TARGETS 1 UP | 194 | 133 | All SZGR 2.0 genes in this pathway |
| KOINUMA TARGETS OF SMAD2 OR SMAD3 | 824 | 528 | All SZGR 2.0 genes in this pathway |