Gene Page: SIPA1L3
Summary ?
| GeneID | 23094 |
| Symbol | SIPA1L3 |
| Synonyms | CTRCT45|SPAL3|SPAR3 |
| Description | signal-induced proliferation-associated 1 like 3 |
| Reference | MIM:616655|HGNC:HGNC:23801|Ensembl:ENSG00000105738|HPRD:18755|Vega:OTTHUMG00000073727 |
| Gene type | protein-coding |
| Map location | 19q13.13 |
| Pascal p-value | 3.149E-4 |
| Sherlock p-value | 0.703 |
| Fetal beta | -0.638 |
| DMG | 1 (# studies) |
| eGene | Myers' cis & trans |
| Support | CompositeSet Darnell FMRP targets |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CV:PGCnp | Genome-wide Association Study | GWAS | |
| DMG:Wockner_2014 | Genome-wide DNA methylation analysis | This dataset includes 4641 differentially methylated probes corresponding to 2929 unique genes between schizophrenia patients (n=24) and controls (n=24). | 1 |
Section I. Genetics and epigenetics annotation
 Differentially methylated gene
Differentially methylated gene
| Probe | Chromosome | Position | Nearest gene | P (dis) | Beta (dis) | FDR (dis) | Study |
|---|---|---|---|---|---|---|---|
| cg24420726 | 19 | 38684319 | SIPA1L3 | 4.594E-4 | 0.707 | 0.046 | DMG:Wockner_2014 |
 eQTL annotation
eQTL annotation
| SNP ID | Chromosome | Position | eGene | Gene Entrez ID | pvalue | qvalue | TSS distance | eQTL type |
|---|---|---|---|---|---|---|---|---|
| snp_a-2071872 | 0 | SIPA1L3 | 23094 | 0.17 | trans | |||
| rs1146358 | chr1 | 116657767 | SIPA1L3 | 23094 | 0 | trans | ||
| rs1934578 | chr1 | 246016476 | SIPA1L3 | 23094 | 0.09 | trans | ||
| rs1934577 | chr1 | 246016542 | SIPA1L3 | 23094 | 0.01 | trans | ||
| rs17012898 | chr2 | 33601648 | SIPA1L3 | 23094 | 0.17 | trans | ||
| rs17012916 | chr2 | 33606031 | SIPA1L3 | 23094 | 0.11 | trans | ||
| rs2716734 | chr2 | 39947720 | SIPA1L3 | 23094 | 2.476E-7 | trans | ||
| rs2716736 | chr2 | 39947946 | SIPA1L3 | 23094 | 2.476E-7 | trans | ||
| rs6543046 | chr2 | 102043302 | SIPA1L3 | 23094 | 0.12 | trans | ||
| rs6754175 | chr2 | 135333296 | SIPA1L3 | 23094 | 0.01 | trans | ||
| rs7651626 | chr3 | 4133729 | SIPA1L3 | 23094 | 0.04 | trans | ||
| rs17039866 | chr3 | 4136543 | SIPA1L3 | 23094 | 0.01 | trans | ||
| rs11928084 | chr3 | 4138812 | SIPA1L3 | 23094 | 5.794E-4 | trans | ||
| rs9882137 | chr3 | 29494146 | SIPA1L3 | 23094 | 0.13 | trans | ||
| rs17052053 | chr3 | 52334592 | SIPA1L3 | 23094 | 0.1 | trans | ||
| rs16850133 | chr3 | 140030776 | SIPA1L3 | 23094 | 2.052E-5 | trans | ||
| rs7638929 | chr3 | 140031075 | SIPA1L3 | 23094 | 1.069E-4 | trans | ||
| rs2597796 | chr4 | 17535859 | SIPA1L3 | 23094 | 0.05 | trans | ||
| rs1248551 | chr4 | 46074678 | SIPA1L3 | 23094 | 0.18 | trans | ||
| rs17023105 | chr4 | 96074164 | SIPA1L3 | 23094 | 0.14 | trans | ||
| rs10516959 | chr4 | 96088848 | SIPA1L3 | 23094 | 0.14 | trans | ||
| rs2713946 | chr4 | 111197804 | SIPA1L3 | 23094 | 0.01 | trans | ||
| rs16869878 | chr5 | 2523760 | SIPA1L3 | 23094 | 9.032E-6 | trans | ||
| rs467856 | chr5 | 3128168 | SIPA1L3 | 23094 | 8.64E-6 | trans | ||
| rs16902549 | chr5 | 13407346 | SIPA1L3 | 23094 | 0.02 | trans | ||
| rs1010527 | chr5 | 13508264 | SIPA1L3 | 23094 | 0.11 | trans | ||
| rs1992711 | chr5 | 13510917 | SIPA1L3 | 23094 | 0.11 | trans | ||
| rs6451640 | chr5 | 20088127 | SIPA1L3 | 23094 | 0.01 | trans | ||
| snp_a-2169623 | 0 | SIPA1L3 | 23094 | 4.063E-6 | trans | |||
| rs6873234 | chr5 | 169299967 | SIPA1L3 | 23094 | 3.234E-4 | trans | ||
| rs7383548 | 0 | SIPA1L3 | 23094 | 3.758E-4 | trans | |||
| rs2016128 | chr6 | 42442743 | SIPA1L3 | 23094 | 0.2 | trans | ||
| rs1587434 | chr6 | 66615354 | SIPA1L3 | 23094 | 3.987E-10 | trans | ||
| rs4557559 | chr6 | 118406801 | SIPA1L3 | 23094 | 0 | trans | ||
| rs6949211 | chr7 | 11231070 | SIPA1L3 | 23094 | 0.17 | trans | ||
| rs17154235 | chr7 | 26692981 | SIPA1L3 | 23094 | 0.07 | trans | ||
| rs4947631 | chr7 | 50603378 | SIPA1L3 | 23094 | 1.938E-4 | trans | ||
| rs10257132 | chr7 | 51422943 | SIPA1L3 | 23094 | 0.01 | trans | ||
| rs17155272 | chr7 | 81248809 | SIPA1L3 | 23094 | 0.15 | trans | ||
| rs529539 | chr7 | 84003500 | SIPA1L3 | 23094 | 0.03 | trans | ||
| rs7796458 | chr7 | 154873701 | SIPA1L3 | 23094 | 0.11 | trans | ||
| rs870686 | chr8 | 25253521 | SIPA1L3 | 23094 | 0.01 | trans | ||
| rs10503771 | chr8 | 25304019 | SIPA1L3 | 23094 | 0.02 | trans | ||
| rs2810511 | chr9 | 136791540 | SIPA1L3 | 23094 | 0.04 | trans | ||
| rs10994470 | chr10 | 51558659 | SIPA1L3 | 23094 | 0.02 | trans | ||
| rs12767540 | chr10 | 56171628 | SIPA1L3 | 23094 | 0.15 | trans | ||
| rs16928875 | chr10 | 73212531 | SIPA1L3 | 23094 | 0.09 | trans | ||
| rs12268485 | chr10 | 83608740 | SIPA1L3 | 23094 | 0.15 | trans | ||
| rs17106311 | chr10 | 87897271 | SIPA1L3 | 23094 | 0.19 | trans | ||
| rs17107175 | chr10 | 88646323 | SIPA1L3 | 23094 | 0.03 | trans | ||
| rs1539587 | chr10 | 115372155 | SIPA1L3 | 23094 | 0.06 | trans | ||
| rs10768704 | chr11 | 41581241 | SIPA1L3 | 23094 | 0.15 | trans | ||
| rs11037691 | chr11 | 43891636 | SIPA1L3 | 23094 | 0.09 | trans | ||
| rs4944475 | chr11 | 83985410 | SIPA1L3 | 23094 | 0.04 | trans | ||
| rs12270706 | chr11 | 109947508 | SIPA1L3 | 23094 | 0.02 | trans | ||
| rs7298298 | chr12 | 17810638 | SIPA1L3 | 23094 | 0.03 | trans | ||
| rs11183748 | chr12 | 47516125 | SIPA1L3 | 23094 | 0.14 | trans | ||
| rs12423193 | chr12 | 63753383 | SIPA1L3 | 23094 | 4.845E-4 | trans | ||
| rs9888446 | chr12 | 63785677 | SIPA1L3 | 23094 | 3.26E-4 | trans | ||
| rs1400878 | chr12 | 83609397 | SIPA1L3 | 23094 | 0 | trans | ||
| rs16945217 | chr12 | 115679171 | SIPA1L3 | 23094 | 0.01 | trans | ||
| rs998923 | chr13 | 35507810 | SIPA1L3 | 23094 | 4.133E-4 | trans | ||
| rs2764585 | chr13 | 46880234 | SIPA1L3 | 23094 | 0.01 | trans | ||
| rs987104 | chr13 | 61340410 | SIPA1L3 | 23094 | 0.2 | trans | ||
| rs9590075 | chr13 | 95414389 | SIPA1L3 | 23094 | 0.1 | trans | ||
| snp_a-2222355 | 0 | SIPA1L3 | 23094 | 0 | trans | |||
| rs7176277 | chr15 | 63316182 | SIPA1L3 | 23094 | 0.05 | trans | ||
| rs6495316 | chr15 | 78986100 | SIPA1L3 | 23094 | 0.11 | trans | ||
| rs1461211 | chr15 | 88628773 | SIPA1L3 | 23094 | 0.05 | trans | ||
| rs16972025 | chr16 | 19461881 | SIPA1L3 | 23094 | 0.03 | trans | ||
| rs1557515 | chr16 | 19462965 | SIPA1L3 | 23094 | 0.17 | trans | ||
| rs4792093 | chr17 | 10939374 | SIPA1L3 | 23094 | 0.02 | trans | ||
| rs7223509 | chr17 | 10940048 | SIPA1L3 | 23094 | 0.02 | trans | ||
| rs1015663 | chr17 | 14238086 | SIPA1L3 | 23094 | 0.01 | trans | ||
| rs295104 | chr17 | 32814665 | SIPA1L3 | 23094 | 0 | trans | ||
| rs349973 | chr17 | 50642288 | SIPA1L3 | 23094 | 0.01 | trans | ||
| rs8078127 | chr17 | 50761342 | SIPA1L3 | 23094 | 0 | trans | ||
| rs10512600 | chr17 | 72812978 | SIPA1L3 | 23094 | 7.026E-4 | trans | ||
| rs2661679 | chr17 | 76172103 | SIPA1L3 | 23094 | 0 | trans | ||
| rs7217417 | chr17 | 76173174 | SIPA1L3 | 23094 | 0 | trans | ||
| rs6040607 | chr20 | 11371092 | SIPA1L3 | 23094 | 0.06 | trans | ||
| rs4270401 | chr20 | 61343887 | SIPA1L3 | 23094 | 2.866E-4 | trans | ||
| rs225434 | chr21 | 43724740 | SIPA1L3 | 23094 | 0.02 | trans | ||
| rs10521889 | chrX | 149836861 | SIPA1L3 | 23094 | 0.03 | trans |
Section II. Transcriptome annotation
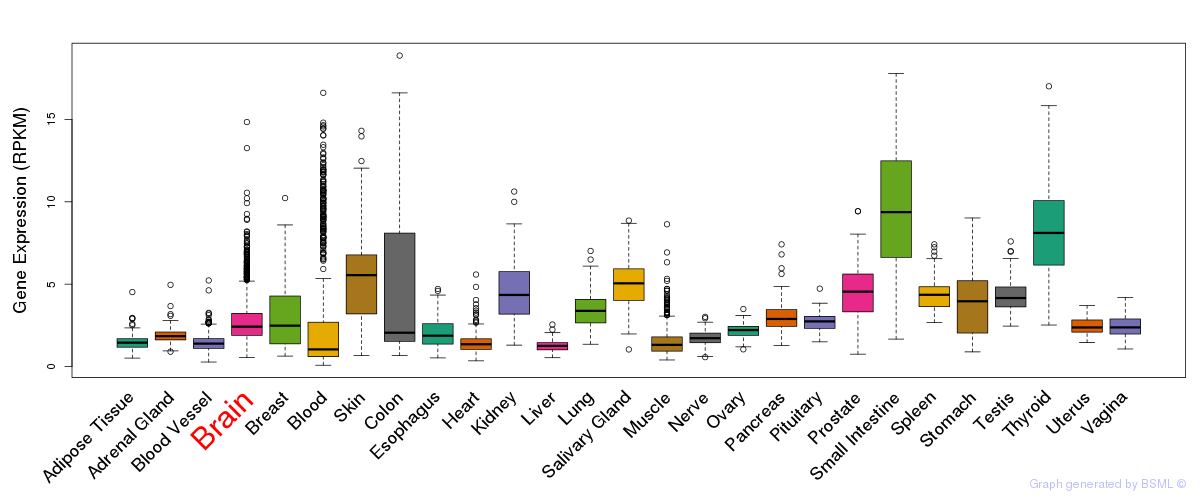
General gene expression (GTEx)
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
Top co-expressed genes in brain regions
| Top 10 positively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| DYRK1A | 0.95 | 0.96 |
| EXOC7 | 0.95 | 0.96 |
| ANKRD17 | 0.94 | 0.95 |
| FAM161B | 0.94 | 0.96 |
| MBD5 | 0.94 | 0.95 |
| TGFBRAP1 | 0.94 | 0.96 |
| ZFYVE1 | 0.94 | 0.95 |
| YTHDF1 | 0.94 | 0.95 |
| N4BP1 | 0.94 | 0.95 |
| KIAA1409 | 0.94 | 0.94 |
| Top 10 negatively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| AF347015.31 | -0.81 | -0.86 |
| MT-CO2 | -0.80 | -0.86 |
| AF347015.21 | -0.78 | -0.90 |
| AF347015.27 | -0.77 | -0.82 |
| HIGD1B | -0.76 | -0.84 |
| AF347015.8 | -0.76 | -0.84 |
| MT-CYB | -0.76 | -0.81 |
| FXYD1 | -0.76 | -0.81 |
| IFI27 | -0.76 | -0.81 |
| AF347015.33 | -0.75 | -0.79 |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| ODONNELL TFRC TARGETS UP | 456 | 228 | All SZGR 2.0 genes in this pathway |
| SENESE HDAC1 AND HDAC2 TARGETS UP | 238 | 144 | All SZGR 2.0 genes in this pathway |
| CHEBOTAEV GR TARGETS DN | 120 | 73 | All SZGR 2.0 genes in this pathway |
| ROVERSI GLIOMA COPY NUMBER UP | 100 | 75 | All SZGR 2.0 genes in this pathway |
| OXFORD RALB TARGETS DN | 9 | 6 | All SZGR 2.0 genes in this pathway |
| FURUKAWA DUSP6 TARGETS PCI35 DN | 74 | 40 | All SZGR 2.0 genes in this pathway |
| NUYTTEN EZH2 TARGETS UP | 1037 | 673 | All SZGR 2.0 genes in this pathway |
| KOYAMA SEMA3B TARGETS UP | 292 | 168 | All SZGR 2.0 genes in this pathway |
| YAGI AML WITH T 8 21 TRANSLOCATION | 368 | 247 | All SZGR 2.0 genes in this pathway |
| MIKKELSEN MCV6 LCP WITH H3K4ME3 | 162 | 80 | All SZGR 2.0 genes in this pathway |
| MIKKELSEN IPS LCP WITH H3K4ME3 | 174 | 100 | All SZGR 2.0 genes in this pathway |
| MIKKELSEN ES LCP WITH H3K4ME3 | 142 | 80 | All SZGR 2.0 genes in this pathway |
| PILON KLF1 TARGETS DN | 1972 | 1213 | All SZGR 2.0 genes in this pathway |