Gene Page: KIF13B
Summary ?
| GeneID | 23303 |
| Symbol | KIF13B |
| Synonyms | GAKIN |
| Description | kinesin family member 13B |
| Reference | MIM:607350|HGNC:HGNC:14405|Ensembl:ENSG00000197892|HPRD:09556|Vega:OTTHUMG00000164032 |
| Gene type | protein-coding |
| Map location | 8p12 |
| Pascal p-value | 0.218 |
| Sherlock p-value | 0.195 |
| Fetal beta | -0.689 |
| eGene | Myers' cis & trans |
| Support | CompositeSet Ascano FMRP targets |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CV:PGCnp | Genome-wide Association Study | GWAS | |
| DNM:Fromer_2014 | Whole Exome Sequencing analysis | This study reported a WES study of 623 schizophrenia trios, reporting DNMs using genomic DNA. | |
| GSMA_IIA | Genome scan meta-analysis (All samples) | Psr: 0.03086 | |
| GSMA_IIE | Genome scan meta-analysis (European-ancestry samples) | Psr: 0.00057 |
Section I. Genetics and epigenetics annotation
 DNM table
DNM table
| Gene | Chromosome | Position | Ref | Alt | Transcript | AA change | Mutation type | Sift | CG46 | Trait | Study |
|---|---|---|---|---|---|---|---|---|---|---|---|
| KIF13B | chr8 | 29025041 | T | C | NM_015254 | p.336N>S | missense | Schizophrenia | DNM:Fromer_2014 |
 eQTL annotation
eQTL annotation
| SNP ID | Chromosome | Position | eGene | Gene Entrez ID | pvalue | qvalue | TSS distance | eQTL type |
|---|---|---|---|---|---|---|---|---|
| rs7830259 | chr8 | 14087643 | KIF13B | 23303 | 0.08 | trans |
Section II. Transcriptome annotation
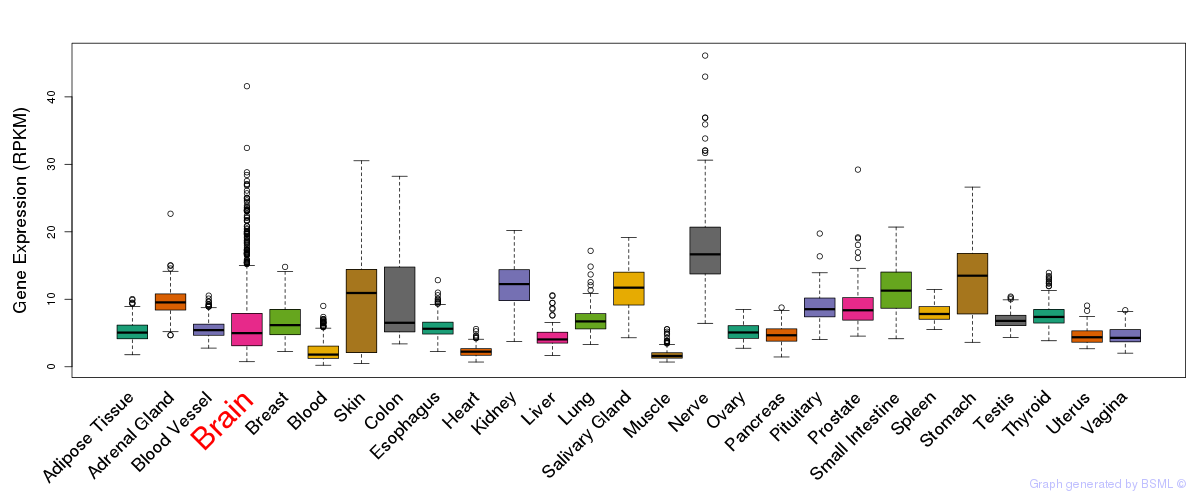
General gene expression (GTEx)
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
Top co-expressed genes in brain regions
| Top 10 positively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| FASTK | 0.77 | 0.80 |
| NDUFAF3 | 0.76 | 0.76 |
| MED29 | 0.76 | 0.75 |
| C19orf50 | 0.75 | 0.74 |
| MPV17 | 0.74 | 0.74 |
| PEX10 | 0.74 | 0.75 |
| ACAA1 | 0.74 | 0.72 |
| C9orf119 | 0.73 | 0.71 |
| COQ9 | 0.72 | 0.72 |
| MRPL17 | 0.72 | 0.70 |
| Top 10 negatively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| EIF5B | -0.45 | -0.51 |
| AC010300.1 | -0.43 | -0.33 |
| SEC62 | -0.42 | -0.45 |
| AF347015.18 | -0.40 | -0.18 |
| AC005921.3 | -0.38 | -0.46 |
| MT-ATP8 | -0.35 | -0.16 |
| EDN1 | -0.35 | -0.17 |
| FAM159B | -0.32 | -0.43 |
| AF347015.21 | -0.32 | -0.13 |
| AP001877.2 | -0.31 | -0.27 |
Section III. Gene Ontology annotation
| Molecular function | GO term | Evidence | Neuro keywords | PubMed ID |
|---|---|---|---|---|
| GO:0000166 | nucleotide binding | IEA | - | |
| GO:0003777 | microtubule motor activity | IEA | - | |
| GO:0003777 | microtubule motor activity | NAS | 10859302 | |
| GO:0005515 | protein binding | IPI | 10859302 | |
| GO:0005524 | ATP binding | IEA | - | |
| GO:0019901 | protein kinase binding | IPI | 10859302 | |
| Biological process | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0007165 | signal transduction | NAS | 10859302 | |
| GO:0007018 | microtubule-based movement | IEA | - | |
| GO:0007018 | microtubule-based movement | TAS | 10859302 | |
| GO:0006605 | protein targeting | TAS | 10859302 | |
| GO:0042110 | T cell activation | NAS | 12496241 | |
| Cellular component | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0005856 | cytoskeleton | IEA | - | |
| GO:0005874 | microtubule | IEA | - | |
| GO:0005875 | microtubule associated complex | IEA | - | |
| GO:0005737 | cytoplasm | IDA | 10859302 |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| PID ARF6 PATHWAY | 35 | 27 | All SZGR 2.0 genes in this pathway |
| SOTIRIOU BREAST CANCER GRADE 1 VS 3 DN | 52 | 34 | All SZGR 2.0 genes in this pathway |
| KINSEY TARGETS OF EWSR1 FLII FUSION UP | 1278 | 748 | All SZGR 2.0 genes in this pathway |
| DODD NASOPHARYNGEAL CARCINOMA UP | 1821 | 933 | All SZGR 2.0 genes in this pathway |
| LUCAS HNF4A TARGETS UP | 58 | 36 | All SZGR 2.0 genes in this pathway |
| RODRIGUES NTN1 AND DCC TARGETS | 35 | 25 | All SZGR 2.0 genes in this pathway |
| MEINHOLD OVARIAN CANCER LOW GRADE UP | 19 | 15 | All SZGR 2.0 genes in this pathway |
| KIM MYC AMPLIFICATION TARGETS DN | 97 | 51 | All SZGR 2.0 genes in this pathway |
| ASTON MAJOR DEPRESSIVE DISORDER DN | 160 | 110 | All SZGR 2.0 genes in this pathway |
| LI WILMS TUMOR VS FETAL KIDNEY 1 UP | 182 | 119 | All SZGR 2.0 genes in this pathway |
| BLALOCK ALZHEIMERS DISEASE UP | 1691 | 1088 | All SZGR 2.0 genes in this pathway |
| CREIGHTON ENDOCRINE THERAPY RESISTANCE 5 | 482 | 296 | All SZGR 2.0 genes in this pathway |
| ACEVEDO LIVER CANCER DN | 540 | 340 | All SZGR 2.0 genes in this pathway |
| SMID BREAST CANCER BASAL DN | 701 | 446 | All SZGR 2.0 genes in this pathway |
| BOQUEST STEM CELL CULTURED VS FRESH UP | 425 | 298 | All SZGR 2.0 genes in this pathway |
| CLIMENT BREAST CANCER COPY NUMBER DN | 8 | 7 | All SZGR 2.0 genes in this pathway |
| VANTVEER BREAST CANCER ESR1 UP | 167 | 99 | All SZGR 2.0 genes in this pathway |
| CAIRO LIVER DEVELOPMENT DN | 222 | 141 | All SZGR 2.0 genes in this pathway |
| ZWANG TRANSIENTLY UP BY 1ST EGF PULSE ONLY | 1839 | 928 | All SZGR 2.0 genes in this pathway |
Section VI. microRNA annotation
| miRNA family | Target position | miRNA ID | miRNA seq | ||
|---|---|---|---|---|---|
| UTR start | UTR end | Match method | |||
| miR-9 | 3086 | 3092 | 1A | hsa-miR-9SZ | UCUUUGGUUAUCUAGCUGUAUGA |
- SZ: miRNAs which differentially expressed in brain cortex of schizophrenia patients comparing with control samples using microarray. Click here to see the list of SZ related miRNAs.
- Brain: miRNAs which are expressed in brain based on miRNA microarray expression studies. Click here to see the list of brain related miRNAs.