Gene Page: RPGRIP1L
Summary ?
| GeneID | 23322 |
| Symbol | RPGRIP1L |
| Synonyms | CORS3|FTM|JBTS7|MKS5|NPHP8|PPP1R134 |
| Description | RPGRIP1-like |
| Reference | MIM:610937|HGNC:HGNC:29168|Ensembl:ENSG00000103494|HPRD:17204|Vega:OTTHUMG00000173125 |
| Gene type | protein-coding |
| Map location | 16q12.2 |
| Pascal p-value | 0.002 |
| Fetal beta | 0.068 |
| eGene | Cortex |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CV:GWASdb | Genome-wide Association Studies | GWASdb records for schizophrenia | |
| CV:PGCnp | Genome-wide Association Study | GWAS | |
| PMID:cooccur | High-throughput literature-search | Systematic search in PubMed for genes co-occurring with SCZ keywords. A total of 3027 genes were included. | |
| Literature | High-throughput literature-search | Co-occurance with Schizophrenia keywords: schizophrenia,schizophrenias | Click to show details |
| GO_Annotation | Mapping neuro-related keywords to Gene Ontology annotations | Hits with neuro-related keywords: 4 |
Section I. Genetics and epigenetics annotation
Section II. Transcriptome annotation
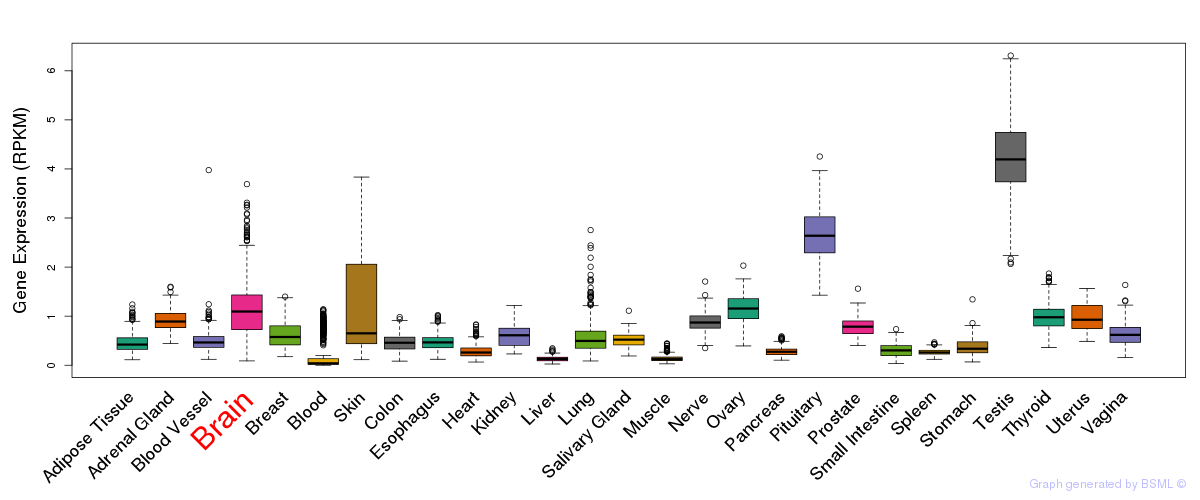
General gene expression (GTEx)
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
Top co-expressed genes in brain regions
| Top 10 positively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| TSN | 0.92 | 0.90 |
| ARMC1 | 0.91 | 0.91 |
| VPS37A | 0.90 | 0.89 |
| UBA2 | 0.90 | 0.93 |
| ELP3 | 0.90 | 0.87 |
| KPNA4 | 0.89 | 0.89 |
| SMU1 | 0.89 | 0.87 |
| CNOT7 | 0.89 | 0.88 |
| RBM18 | 0.89 | 0.89 |
| MAPKSP1 | 0.89 | 0.87 |
| Top 10 negatively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| MT-CO2 | -0.75 | -0.75 |
| AF347015.8 | -0.73 | -0.75 |
| MT-CYB | -0.72 | -0.74 |
| AF347015.33 | -0.71 | -0.72 |
| AF347015.31 | -0.71 | -0.73 |
| AF347015.21 | -0.70 | -0.72 |
| AF347015.26 | -0.70 | -0.72 |
| AF347015.2 | -0.69 | -0.70 |
| AF347015.15 | -0.69 | -0.73 |
| AF347015.27 | -0.68 | -0.71 |
Section III. Gene Ontology annotation
| Biological process | GO term | Evidence | Neuro keywords | PubMed ID |
|---|---|---|---|---|
| GO:0007420 | brain development | IEA | Brain (GO term level: 7) | - |
| GO:0022038 | corpus callosum development | IEA | axon (GO term level: 10) | - |
| GO:0021549 | cerebellum development | IEA | Brain (GO term level: 10) | - |
| GO:0021772 | olfactory bulb development | IEA | neuron (GO term level: 11) | - |
| GO:0001701 | in utero embryonic development | IEA | - | |
| GO:0001889 | liver development | IEA | - | |
| GO:0001822 | kidney development | IEA | - | |
| GO:0007368 | determination of left/right symmetry | IEA | - | |
| GO:0008589 | regulation of smoothened signaling pathway | IEA | - | |
| GO:0007382 | specification of segmental identity, maxillary segment | IEA | - | |
| GO:0021532 | neural tube patterning | IEA | - | |
| GO:0042305 | specification of segmental identity, mandibular segment | IEA | - | |
| GO:0042384 | cilium biogenesis | IEA | - | |
| GO:0021670 | lateral ventricle development | IEA | - | |
| GO:0043010 | camera-type eye development | IEA | - | |
| GO:0035108 | limb morphogenesis | IEA | - | |
| GO:0035115 | embryonic forelimb morphogenesis | IEA | - | |
| GO:0035116 | embryonic hindlimb morphogenesis | IEA | - | |
| GO:0043584 | nose development | IEA | - | |
| GO:0060039 | pericardium development | IEA | - | |
| Cellular component | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0005813 | centrosome | IDA | 17558409 | |
| GO:0005737 | cytoplasm | IDA | 17558409 | |
| GO:0005932 | basal body | IDA | 17558409 | |
| GO:0005929 | cilium | IDA | 17558409 | |
| GO:0035085 | cilium axoneme | IDA | 17558409 |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| ROYLANCE BREAST CANCER 16Q COPY NUMBER UP | 63 | 44 | All SZGR 2.0 genes in this pathway |
| GEORGES TARGETS OF MIR192 AND MIR215 | 893 | 528 | All SZGR 2.0 genes in this pathway |
| KRIGE RESPONSE TO TOSEDOSTAT 6HR DN | 911 | 527 | All SZGR 2.0 genes in this pathway |
| KRIGE RESPONSE TO TOSEDOSTAT 24HR DN | 1011 | 592 | All SZGR 2.0 genes in this pathway |
| CREIGHTON ENDOCRINE THERAPY RESISTANCE 2 | 473 | 224 | All SZGR 2.0 genes in this pathway |
| MITSIADES RESPONSE TO APLIDIN UP | 439 | 257 | All SZGR 2.0 genes in this pathway |
Section VI. microRNA annotation
| miRNA family | Target position | miRNA ID | miRNA seq | ||
|---|---|---|---|---|---|
| UTR start | UTR end | Match method | |||
| miR-103/107 | 1008 | 1015 | 1A,m8 | hsa-miR-103brain | AGCAGCAUUGUACAGGGCUAUGA |
| hsa-miR-107brain | AGCAGCAUUGUACAGGGCUAUCA | ||||
| miR-125/351 | 1166 | 1172 | 1A | hsa-miR-125bbrain | UCCCUGAGACCCUAACUUGUGA |
| hsa-miR-125abrain | UCCCUGAGACCCUUUAACCUGUG | ||||
| miR-128 | 453 | 459 | 1A | hsa-miR-128a | UCACAGUGAACCGGUCUCUUUU |
| hsa-miR-128b | UCACAGUGAACCGGUCUCUUUC | ||||
| miR-133 | 1540 | 1546 | m8 | hsa-miR-133a | UUGGUCCCCUUCAACCAGCUGU |
| hsa-miR-133b | UUGGUCCCCUUCAACCAGCUA | ||||
| miR-136 | 991 | 997 | m8 | hsa-miR-136 | ACUCCAUUUGUUUUGAUGAUGGA |
| miR-137 | 472 | 479 | 1A,m8 | hsa-miR-137 | UAUUGCUUAAGAAUACGCGUAG |
| miR-144 | 451 | 457 | m8 | hsa-miR-144 | UACAGUAUAGAUGAUGUACUAG |
| miR-185 | 1721 | 1727 | 1A | hsa-miR-185brain | UGGAGAGAAAGGCAGUUC |
| miR-196 | 1193 | 1199 | 1A | hsa-miR-196a | UAGGUAGUUUCAUGUUGUUGG |
| hsa-miR-196b | UAGGUAGUUUCCUGUUGUUGG | ||||
| miR-203.1 | 1049 | 1055 | 1A | hsa-miR-203 | UGAAAUGUUUAGGACCACUAG |
| hsa-miR-203 | UGAAAUGUUUAGGACCACUAG | ||||
| miR-216 | 62 | 68 | m8 | hsa-miR-216 | UAAUCUCAGCUGGCAACUGUG |
| miR-217 | 1853 | 1859 | 1A | hsa-miR-217 | UACUGCAUCAGGAACUGAUUGGAU |
| miR-22 | 1398 | 1404 | 1A | hsa-miR-22brain | AAGCUGCCAGUUGAAGAACUGU |
| miR-27 | 998 | 1004 | 1A | hsa-miR-27abrain | UUCACAGUGGCUAAGUUCCGC |
| hsa-miR-27bbrain | UUCACAGUGGCUAAGUUCUGC | ||||
| hsa-miR-27abrain | UUCACAGUGGCUAAGUUCCGC | ||||
| hsa-miR-27bbrain | UUCACAGUGGCUAAGUUCUGC | ||||
| miR-28 | 1308 | 1314 | 1A | hsa-miR-28brain | AAGGAGCUCACAGUCUAUUGAG |
| miR-323 | 498 | 504 | m8 | hsa-miR-323brain | GCACAUUACACGGUCGACCUCU |
| miR-34/449 | 197 | 203 | 1A | hsa-miR-34abrain | UGGCAGUGUCUUAGCUGGUUGUU |
| hsa-miR-34c | AGGCAGUGUAGUUAGCUGAUUGC | ||||
| hsa-miR-449 | UGGCAGUGUAUUGUUAGCUGGU | ||||
| hsa-miR-449b | AGGCAGUGUAUUGUUAGCUGGC | ||||
| miR-377 | 1275 | 1281 | m8 | hsa-miR-377 | AUCACACAAAGGCAACUUUUGU |
| miR-381 | 150 | 156 | 1A | hsa-miR-381 | UAUACAAGGGCAAGCUCUCUGU |
| miR-409-3p | 1321 | 1328 | 1A,m8 | hsa-miR-409-3p | CGAAUGUUGCUCGGUGAACCCCU |
| miR-452 | 1125 | 1132 | 1A,m8 | hsa-miR-452 | UGUUUGCAGAGGAAACUGAGAC |
| miR-488 | 900 | 906 | m8 | hsa-miR-488 | CCCAGAUAAUGGCACUCUCAA |
| miR-495 | 1748 | 1754 | m8 | hsa-miR-495brain | AAACAAACAUGGUGCACUUCUUU |
| miR-496 | 1155 | 1161 | 1A | hsa-miR-496 | AUUACAUGGCCAAUCUC |
- SZ: miRNAs which differentially expressed in brain cortex of schizophrenia patients comparing with control samples using microarray. Click here to see the list of SZ related miRNAs.
- Brain: miRNAs which are expressed in brain based on miRNA microarray expression studies. Click here to see the list of brain related miRNAs.