Gene Page: GABRA2
Summary ?
| GeneID | 2555 |
| Symbol | GABRA2 |
| Synonyms | - |
| Description | gamma-aminobutyric acid type A receptor alpha2 subunit |
| Reference | MIM:137140|HGNC:HGNC:4076|Ensembl:ENSG00000151834|HPRD:11747|Vega:OTTHUMG00000044266 |
| Gene type | protein-coding |
| Map location | 4p12 |
| Pascal p-value | 8.798E-4 |
| Sherlock p-value | 0.842 |
| Fetal beta | -1.169 |
| eGene | Myers' cis & trans |
| Support | LIGAND GATED ION SIGNALING |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CV:PGCnp | Genome-wide Association Study | GWAS | |
| PMID:cooccur | High-throughput literature-search | Systematic search in PubMed for genes co-occurring with SCZ keywords. A total of 3027 genes were included. | |
| Literature | High-throughput literature-search | Co-occurance with Schizophrenia keywords: schizophrenia,schizophrenic,schizophrenias | Click to show details |
| GO_Annotation | Mapping neuro-related keywords to Gene Ontology annotations | Hits with neuro-related keywords: 7 |
Section I. Genetics and epigenetics annotation
 eQTL annotation
eQTL annotation
| SNP ID | Chromosome | Position | eGene | Gene Entrez ID | pvalue | qvalue | TSS distance | eQTL type |
|---|---|---|---|---|---|---|---|---|
| rs17145698 | chrX | 40218345 | GABRA2 | 2555 | 0.16 | trans |
Section II. Transcriptome annotation
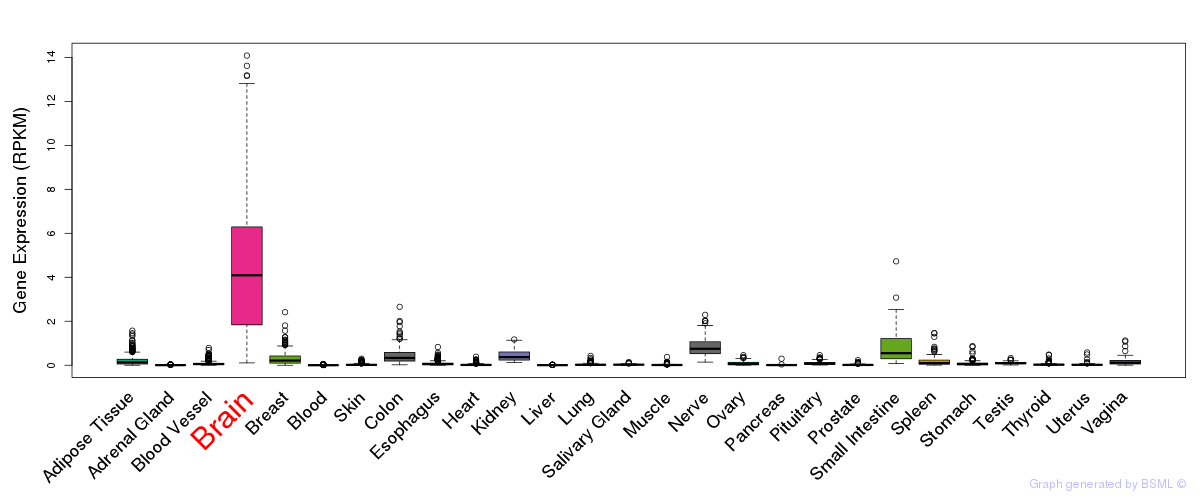
General gene expression (GTEx)
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
Top co-expressed genes in brain regions
| Top 10 positively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| NRXN3 | 0.83 | 0.93 |
| OXCT1 | 0.83 | 0.88 |
| PARK2 | 0.82 | 0.87 |
| PRMT8 | 0.82 | 0.85 |
| NPTN | 0.81 | 0.84 |
| MAPK13 | 0.81 | 0.83 |
| ATP6AP2 | 0.81 | 0.84 |
| RASGEF1A | 0.80 | 0.85 |
| DRP2 | 0.80 | 0.87 |
| ZADH2 | 0.80 | 0.84 |
| Top 10 negatively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| AF347015.21 | -0.40 | -0.24 |
| AF347015.18 | -0.39 | -0.26 |
| AF347015.2 | -0.35 | -0.21 |
| RPL38 | -0.34 | -0.40 |
| ANP32C | -0.34 | -0.34 |
| EMX2 | -0.34 | -0.42 |
| FAM159B | -0.33 | -0.47 |
| MT-CO2 | -0.33 | -0.24 |
| AL139819.3 | -0.33 | -0.29 |
| AF347015.26 | -0.33 | -0.18 |
Section III. Gene Ontology annotation
| Molecular function | GO term | Evidence | Neuro keywords | PubMed ID |
|---|---|---|---|---|
| GO:0004890 | GABA-A receptor activity | IEA | GABA (GO term level: 7) | - |
| GO:0008503 | benzodiazepine receptor activity | IMP | GABA, Neurotransmitter (GO term level: 6) | 8391122 |
| GO:0005254 | chloride channel activity | IEA | - | |
| GO:0005216 | ion channel activity | IEA | - | |
| GO:0005230 | extracellular ligand-gated ion channel activity | IEA | - | |
| GO:0031404 | chloride ion binding | IEA | - | |
| Biological process | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0007214 | gamma-aminobutyric acid signaling pathway | IMP | GABA, Neurotransmitter (GO term level: 8) | 8391122 |
| GO:0006836 | neurotransmitter transport | IMP | neuron, Neurotransmitter (GO term level: 5) | 8391122 |
| GO:0001505 | regulation of neurotransmitter levels | IMP | Synap, Neurotransmitter (GO term level: 7) | 8391122 |
| GO:0006821 | chloride transport | IEA | - | |
| GO:0006811 | ion transport | IEA | - | |
| Cellular component | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0030285 | integral to synaptic vesicle membrane | IDA | Synap (GO term level: 14) | 8391122 |
| GO:0045211 | postsynaptic membrane | IEA | Synap, Neurotransmitter (GO term level: 5) | - |
| GO:0045202 | synapse | IEA | neuron, Synap, Neurotransmitter, Glial (GO term level: 2) | - |
| GO:0005886 | plasma membrane | IEA | - | |
| GO:0030054 | cell junction | IEA | - |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| KEGG NEUROACTIVE LIGAND RECEPTOR INTERACTION | 272 | 195 | All SZGR 2.0 genes in this pathway |
| BIOCARTA GABA PATHWAY | 10 | 10 | All SZGR 2.0 genes in this pathway |
| REACTOME TRANSMISSION ACROSS CHEMICAL SYNAPSES | 186 | 155 | All SZGR 2.0 genes in this pathway |
| REACTOME NEURONAL SYSTEM | 279 | 221 | All SZGR 2.0 genes in this pathway |
| REACTOME NEUROTRANSMITTER RECEPTOR BINDING AND DOWNSTREAM TRANSMISSION IN THE POSTSYNAPTIC CELL | 137 | 110 | All SZGR 2.0 genes in this pathway |
| REACTOME TRANSMEMBRANE TRANSPORT OF SMALL MOLECULES | 413 | 270 | All SZGR 2.0 genes in this pathway |
| REACTOME GABA A RECEPTOR ACTIVATION | 12 | 12 | All SZGR 2.0 genes in this pathway |
| REACTOME GABA RECEPTOR ACTIVATION | 52 | 40 | All SZGR 2.0 genes in this pathway |
| REACTOME ION CHANNEL TRANSPORT | 55 | 42 | All SZGR 2.0 genes in this pathway |
| REACTOME LIGAND GATED ION CHANNEL TRANSPORT | 21 | 21 | All SZGR 2.0 genes in this pathway |
| HATADA METHYLATED IN LUNG CANCER UP | 390 | 236 | All SZGR 2.0 genes in this pathway |
| BENPORATH SUZ12 TARGETS | 1038 | 678 | All SZGR 2.0 genes in this pathway |
| BENPORATH EED TARGETS | 1062 | 725 | All SZGR 2.0 genes in this pathway |
| BENPORATH ES WITH H3K27ME3 | 1118 | 744 | All SZGR 2.0 genes in this pathway |
| BENPORATH PRC2 TARGETS | 652 | 441 | All SZGR 2.0 genes in this pathway |
| LEE CALORIE RESTRICTION NEOCORTEX UP | 83 | 66 | All SZGR 2.0 genes in this pathway |
| BLALOCK ALZHEIMERS DISEASE DN | 1237 | 837 | All SZGR 2.0 genes in this pathway |
| BANDRES RESPONSE TO CARMUSTIN MGMT 24HR DN | 33 | 21 | All SZGR 2.0 genes in this pathway |
| BANDRES RESPONSE TO CARMUSTIN MGMT 48HR DN | 161 | 105 | All SZGR 2.0 genes in this pathway |
| WANG SMARCE1 TARGETS UP | 280 | 183 | All SZGR 2.0 genes in this pathway |
| YAUCH HEDGEHOG SIGNALING PARACRINE DN | 264 | 159 | All SZGR 2.0 genes in this pathway |
| HAN SATB1 TARGETS DN | 442 | 275 | All SZGR 2.0 genes in this pathway |
| KYNG WERNER SYNDROM AND NORMAL AGING UP | 93 | 62 | All SZGR 2.0 genes in this pathway |
| KIM ALL DISORDERS CALB1 CORR UP | 548 | 370 | All SZGR 2.0 genes in this pathway |
| BRUINS UVC RESPONSE VIA TP53 GROUP A | 898 | 516 | All SZGR 2.0 genes in this pathway |
| PURBEY TARGETS OF CTBP1 NOT SATB1 DN | 448 | 282 | All SZGR 2.0 genes in this pathway |
| PHONG TNF RESPONSE VIA P38 COMPLETE | 227 | 151 | All SZGR 2.0 genes in this pathway |
| PHONG TNF RESPONSE VIA P38 PARTIAL | 160 | 106 | All SZGR 2.0 genes in this pathway |