Gene Page: GABRP
Summary ?
| GeneID | 2568 |
| Symbol | GABRP |
| Synonyms | - |
| Description | gamma-aminobutyric acid type A receptor pi subunit |
| Reference | MIM:602729|HGNC:HGNC:4089|Ensembl:ENSG00000094755|HPRD:04105|Vega:OTTHUMG00000130443 |
| Gene type | protein-coding |
| Map location | 5q35.1 |
| Pascal p-value | 0.655 |
| Fetal beta | -0.321 |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CV:PGCnp | Genome-wide Association Study | GWAS | |
| PMID:cooccur | High-throughput literature-search | Systematic search in PubMed for genes co-occurring with SCZ keywords. A total of 3027 genes were included. | |
| Association | A combined odds ratio method (Sun et al. 2008), association studies | 2 | Link to SZGene |
| GSMA_IIA | Genome scan meta-analysis (All samples) | Psr: 0.0276 | |
| Literature | High-throughput literature-search | Co-occurance with Schizophrenia keywords: schizophrenia,schizophrenic,schizophrenias | Click to show details |
| GO_Annotation | Mapping neuro-related keywords to Gene Ontology annotations | Hits with neuro-related keywords: 3 |
Section I. Genetics and epigenetics annotation
Section II. Transcriptome annotation
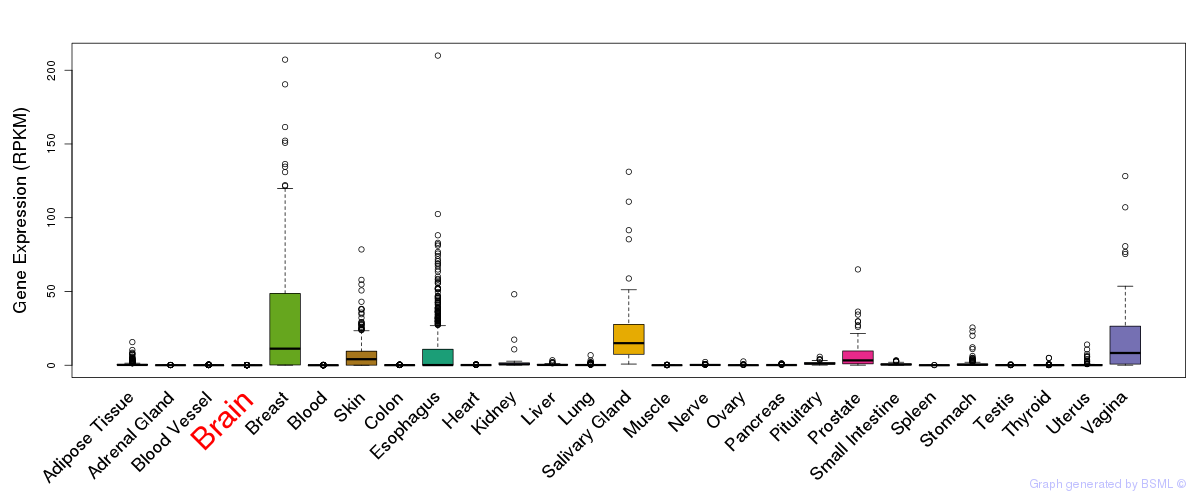
General gene expression (GTEx)
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
Top co-expressed genes in brain regions
| Top 10 positively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| WDR34 | 0.83 | 0.80 |
| ADCK4 | 0.82 | 0.80 |
| B9D2 | 0.81 | 0.76 |
| GSTP1 | 0.81 | 0.81 |
| FAM125A | 0.80 | 0.78 |
| C19orf48 | 0.79 | 0.75 |
| POLD2 | 0.79 | 0.76 |
| FUZ | 0.79 | 0.78 |
| HEMK1 | 0.79 | 0.74 |
| MAD2L2 | 0.79 | 0.75 |
| Top 10 negatively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| AF347015.27 | -0.47 | -0.54 |
| MT-CYB | -0.46 | -0.55 |
| AF347015.15 | -0.45 | -0.54 |
| AF347015.8 | -0.45 | -0.53 |
| MT-CO2 | -0.45 | -0.51 |
| AF347015.26 | -0.44 | -0.55 |
| AF347015.33 | -0.44 | -0.51 |
| AF347015.31 | -0.44 | -0.50 |
| AF347015.2 | -0.43 | -0.51 |
| MT-ATP8 | -0.42 | -0.57 |
Section III. Gene Ontology annotation
| Molecular function | GO term | Evidence | Neuro keywords | PubMed ID |
|---|---|---|---|---|
| GO:0030594 | neurotransmitter receptor activity | IEA | Neurotransmitter (GO term level: 5) | - |
| GO:0004890 | GABA-A receptor activity | TAS | GABA (GO term level: 7) | 9182563 |
| GO:0005254 | chloride channel activity | IEA | - | |
| GO:0005216 | ion channel activity | IEA | - | |
| GO:0005230 | extracellular ligand-gated ion channel activity | IEA | - | |
| GO:0031404 | chloride ion binding | IEA | - | |
| Biological process | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0006811 | ion transport | IEA | - | |
| Cellular component | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0045211 | postsynaptic membrane | IEA | Synap, Neurotransmitter (GO term level: 5) | - |
| GO:0045202 | synapse | IEA | neuron, Synap, Neurotransmitter, Glial (GO term level: 2) | - |
| GO:0016021 | integral to membrane | IEA | - | |
| GO:0005886 | plasma membrane | IEA | - | |
| GO:0030054 | cell junction | IEA | - |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| KEGG NEUROACTIVE LIGAND RECEPTOR INTERACTION | 272 | 195 | All SZGR 2.0 genes in this pathway |
| SENGUPTA NASOPHARYNGEAL CARCINOMA WITH LMP1 DN | 175 | 82 | All SZGR 2.0 genes in this pathway |
| TURASHVILI BREAST NORMAL DUCTAL VS LOBULAR UP | 68 | 40 | All SZGR 2.0 genes in this pathway |
| TURASHVILI BREAST DUCTAL CARCINOMA VS DUCTAL NORMAL DN | 198 | 110 | All SZGR 2.0 genes in this pathway |
| TURASHVILI BREAST DUCTAL CARCINOMA VS LOBULAR NORMAL DN | 69 | 43 | All SZGR 2.0 genes in this pathway |
| TURASHVILI BREAST LOBULAR CARCINOMA VS DUCTAL NORMAL DN | 91 | 53 | All SZGR 2.0 genes in this pathway |
| TURASHVILI BREAST LOBULAR CARCINOMA VS LOBULAR NORMAL UP | 94 | 59 | All SZGR 2.0 genes in this pathway |
| DOANE BREAST CANCER ESR1 DN | 48 | 28 | All SZGR 2.0 genes in this pathway |
| ROY WOUND BLOOD VESSEL DN | 22 | 14 | All SZGR 2.0 genes in this pathway |
| SABATES COLORECTAL ADENOMA UP | 141 | 75 | All SZGR 2.0 genes in this pathway |
| DODD NASOPHARYNGEAL CARCINOMA UP | 1821 | 933 | All SZGR 2.0 genes in this pathway |
| MCBRYAN PUBERTAL BREAST 3 4WK UP | 214 | 144 | All SZGR 2.0 genes in this pathway |
| FARMER BREAST CANCER APOCRINE VS BASAL | 330 | 217 | All SZGR 2.0 genes in this pathway |
| FARMER BREAST CANCER BASAL VS LULMINAL | 330 | 215 | All SZGR 2.0 genes in this pathway |
| HEIDENBLAD AMPLICON 12P11 12 DN | 30 | 16 | All SZGR 2.0 genes in this pathway |
| YANG BREAST CANCER ESR1 DN | 25 | 15 | All SZGR 2.0 genes in this pathway |
| RICKMAN HEAD AND NECK CANCER A | 100 | 63 | All SZGR 2.0 genes in this pathway |
| LIN SILENCED BY TUMOR MICROENVIRONMENT | 108 | 73 | All SZGR 2.0 genes in this pathway |
| MANALO HYPOXIA UP | 207 | 145 | All SZGR 2.0 genes in this pathway |
| JAZAERI BREAST CANCER BRCA1 VS BRCA2 DN | 43 | 31 | All SZGR 2.0 genes in this pathway |
| RODWELL AGING KIDNEY NO BLOOD UP | 222 | 139 | All SZGR 2.0 genes in this pathway |
| RODWELL AGING KIDNEY UP | 487 | 303 | All SZGR 2.0 genes in this pathway |
| HEDENFALK BREAST CANCER BRCA1 VS BRCA2 | 163 | 113 | All SZGR 2.0 genes in this pathway |
| MARTINEZ RB1 TARGETS UP | 673 | 430 | All SZGR 2.0 genes in this pathway |
| MARTINEZ TP53 TARGETS DN | 593 | 372 | All SZGR 2.0 genes in this pathway |
| MARTINEZ RB1 AND TP53 TARGETS DN | 591 | 366 | All SZGR 2.0 genes in this pathway |
| IWANAGA CARCINOGENESIS BY KRAS PTEN UP | 181 | 112 | All SZGR 2.0 genes in this pathway |
| RIZKI TUMOR INVASIVENESS 2D DN | 64 | 35 | All SZGR 2.0 genes in this pathway |
| SMID BREAST CANCER RELAPSE IN BONE DN | 315 | 197 | All SZGR 2.0 genes in this pathway |
| SMID BREAST CANCER LUMINAL B DN | 564 | 326 | All SZGR 2.0 genes in this pathway |
| SMID BREAST CANCER ERBB2 DN | 5 | 5 | All SZGR 2.0 genes in this pathway |
| SMID BREAST CANCER BASAL UP | 648 | 398 | All SZGR 2.0 genes in this pathway |
| HUPER BREAST BASAL VS LUMINAL DN | 59 | 44 | All SZGR 2.0 genes in this pathway |
| VANTVEER BREAST CANCER ESR1 DN | 240 | 153 | All SZGR 2.0 genes in this pathway |
| LEE BMP2 TARGETS UP | 745 | 475 | All SZGR 2.0 genes in this pathway |
| BOSCO EPITHELIAL DIFFERENTIATION MODULE | 69 | 31 | All SZGR 2.0 genes in this pathway |
| LIM MAMMARY STEM CELL DN | 428 | 246 | All SZGR 2.0 genes in this pathway |