Gene Page: SETBP1
Summary ?
| GeneID | 26040 |
| Symbol | SETBP1 |
| Synonyms | MRD29|SEB |
| Description | SET binding protein 1 |
| Reference | MIM:611060|HGNC:HGNC:15573|Ensembl:ENSG00000152217|HPRD:10224|Vega:OTTHUMG00000132613 |
| Gene type | protein-coding |
| Map location | 18q21.1 |
| Pascal p-value | 0.296 |
| Sherlock p-value | 0.309 |
| Fetal beta | 2.515 |
| DMG | 1 (# studies) |
| eGene | Myers' cis & trans |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CV:PGCnp | Genome-wide Association Study | GWAS | |
| DMG:Wockner_2014 | Genome-wide DNA methylation analysis | This dataset includes 4641 differentially methylated probes corresponding to 2929 unique genes between schizophrenia patients (n=24) and controls (n=24). | 1 |
Section I. Genetics and epigenetics annotation
 Differentially methylated gene
Differentially methylated gene
| Probe | Chromosome | Position | Nearest gene | P (dis) | Beta (dis) | FDR (dis) | Study |
|---|---|---|---|---|---|---|---|
| cg24983248 | 18 | 42644213 | SETBP1 | 5.573E-4 | 0.401 | 0.049 | DMG:Wockner_2014 |
 eQTL annotation
eQTL annotation
| SNP ID | Chromosome | Position | eGene | Gene Entrez ID | pvalue | qvalue | TSS distance | eQTL type |
|---|---|---|---|---|---|---|---|---|
| rs1011531 | chr9 | 13755191 | SETBP1 | 26040 | 0.14 | trans |
Section II. Transcriptome annotation
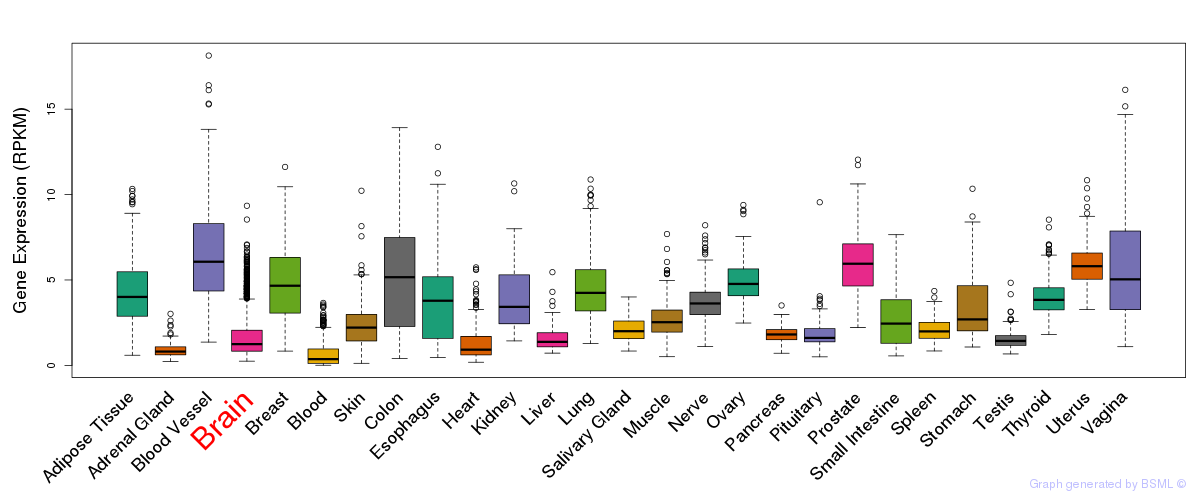
General gene expression (GTEx)
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
Top co-expressed genes in brain regions
| Top 10 positively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| C11orf70 | 0.85 | 0.50 |
| DNALI1 | 0.83 | 0.47 |
| AKAP14 | 0.82 | 0.40 |
| FOXJ1 | 0.81 | 0.34 |
| C6orf118 | 0.81 | 0.37 |
| RIBC2 | 0.81 | 0.38 |
| ARMC4 | 0.81 | 0.37 |
| C19orf51 | 0.81 | 0.32 |
| MORN5 | 0.80 | 0.25 |
| C6orf165 | 0.80 | 0.39 |
| Top 10 negatively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| MGLL | -0.25 | -0.27 |
| MUC5B | -0.24 | -0.35 |
| PK4P | -0.24 | -0.30 |
| BCL2L2 | -0.24 | -0.29 |
| AF347015.15 | -0.24 | -0.31 |
| HR | -0.23 | -0.33 |
| AF347015.2 | -0.23 | -0.30 |
| CABLES1 | -0.23 | -0.31 |
| ARRDC2 | -0.22 | -0.34 |
| AF347015.8 | -0.22 | -0.26 |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| CORRE MULTIPLE MYELOMA DN | 62 | 41 | All SZGR 2.0 genes in this pathway |
| VECCHI GASTRIC CANCER EARLY DN | 367 | 220 | All SZGR 2.0 genes in this pathway |
| HOEBEKE LYMPHOID STEM CELL UP | 95 | 64 | All SZGR 2.0 genes in this pathway |
| SABATES COLORECTAL ADENOMA DN | 291 | 176 | All SZGR 2.0 genes in this pathway |
| DELYS THYROID CANCER DN | 232 | 154 | All SZGR 2.0 genes in this pathway |
| NUYTTEN NIPP1 TARGETS UP | 769 | 437 | All SZGR 2.0 genes in this pathway |
| NUYTTEN EZH2 TARGETS UP | 1037 | 673 | All SZGR 2.0 genes in this pathway |
| SCHAEFFER PROSTATE DEVELOPMENT 12HR UP | 116 | 79 | All SZGR 2.0 genes in this pathway |
| NOUZOVA METHYLATED IN APL | 68 | 39 | All SZGR 2.0 genes in this pathway |
| IVANOVA HEMATOPOIESIS STEM CELL LONG TERM | 302 | 191 | All SZGR 2.0 genes in this pathway |
| HELLER HDAC TARGETS DN | 292 | 189 | All SZGR 2.0 genes in this pathway |
| HELLER HDAC TARGETS SILENCED BY METHYLATION DN | 281 | 179 | All SZGR 2.0 genes in this pathway |
| BOQUEST STEM CELL UP | 260 | 174 | All SZGR 2.0 genes in this pathway |
| SHAFFER IRF4 TARGETS IN ACTIVATED DENDRITIC CELL | 65 | 49 | All SZGR 2.0 genes in this pathway |
| VALK AML CLUSTER 10 | 33 | 22 | All SZGR 2.0 genes in this pathway |
| KOINUMA TARGETS OF SMAD2 OR SMAD3 | 824 | 528 | All SZGR 2.0 genes in this pathway |
| FORTSCHEGGER PHF8 TARGETS DN | 784 | 464 | All SZGR 2.0 genes in this pathway |
| PEDRIOLI MIR31 TARGETS DN | 418 | 245 | All SZGR 2.0 genes in this pathway |
| GENTLES LEUKEMIC STEM CELL UP | 29 | 15 | All SZGR 2.0 genes in this pathway |
| ZWANG TRANSIENTLY UP BY 1ST EGF PULSE ONLY | 1839 | 928 | All SZGR 2.0 genes in this pathway |