| KEGG STARCH AND SUCROSE METABOLISM
| 52 | 33 | All SZGR 2.0 genes in this pathway |
| REACTOME METABOLISM OF CARBOHYDRATES
| 247 | 154 | All SZGR 2.0 genes in this pathway |
| REACTOME GLUCOSE METABOLISM
| 69 | 42 | All SZGR 2.0 genes in this pathway |
| FRASOR TAMOXIFEN RESPONSE UP
| 51 | 36 | All SZGR 2.0 genes in this pathway |
| GARY CD5 TARGETS UP
| 473 | 314 | All SZGR 2.0 genes in this pathway |
| DIAZ CHRONIC MEYLOGENOUS LEUKEMIA UP
| 1382 | 904 | All SZGR 2.0 genes in this pathway |
| BASAKI YBX1 TARGETS DN
| 384 | 230 | All SZGR 2.0 genes in this pathway |
| OSMAN BLADDER CANCER UP
| 404 | 246 | All SZGR 2.0 genes in this pathway |
| RHEIN ALL GLUCOCORTICOID THERAPY DN
| 362 | 238 | All SZGR 2.0 genes in this pathway |
| ELVIDGE HYPOXIA UP
| 171 | 112 | All SZGR 2.0 genes in this pathway |
| ELVIDGE HYPOXIA BY DMOG UP
| 130 | 85 | All SZGR 2.0 genes in this pathway |
| ELVIDGE HIF1A TARGETS DN
| 91 | 58 | All SZGR 2.0 genes in this pathway |
| ELVIDGE HIF1A AND HIF2A TARGETS DN
| 104 | 72 | All SZGR 2.0 genes in this pathway |
| ENK UV RESPONSE KERATINOCYTE DN
| 485 | 334 | All SZGR 2.0 genes in this pathway |
| GRAESSMANN APOPTOSIS BY SERUM DEPRIVATION DN
| 234 | 147 | All SZGR 2.0 genes in this pathway |
| LANDIS ERBB2 BREAST PRENEOPLASTIC DN
| 55 | 33 | All SZGR 2.0 genes in this pathway |
| GRAESSMANN RESPONSE TO MC AND SERUM DEPRIVATION DN
| 84 | 54 | All SZGR 2.0 genes in this pathway |
| BERENJENO TRANSFORMED BY RHOA FOREVER UP
| 19 | 13 | All SZGR 2.0 genes in this pathway |
| LANDIS ERBB2 BREAST TUMORS 324 DN
| 149 | 93 | All SZGR 2.0 genes in this pathway |
| BERENJENO TRANSFORMED BY RHOA UP
| 536 | 340 | All SZGR 2.0 genes in this pathway |
| HERNANDEZ MITOTIC ARREST BY DOCETAXEL 1 DN
| 38 | 23 | All SZGR 2.0 genes in this pathway |
| DACOSTA UV RESPONSE VIA ERCC3 DN
| 855 | 609 | All SZGR 2.0 genes in this pathway |
| DACOSTA UV RESPONSE VIA ERCC3 COMMON DN
| 483 | 336 | All SZGR 2.0 genes in this pathway |
| MARTORIATI MDM4 TARGETS FETAL LIVER UP
| 227 | 137 | All SZGR 2.0 genes in this pathway |
| MAINA VHL TARGETS DN
| 18 | 14 | All SZGR 2.0 genes in this pathway |
| GROSS ELK3 TARGETS DN
| 32 | 17 | All SZGR 2.0 genes in this pathway |
| GROSS HYPOXIA VIA ELK3 DN
| 156 | 106 | All SZGR 2.0 genes in this pathway |
| GROSS HIF1A TARGETS DN
| 25 | 15 | All SZGR 2.0 genes in this pathway |
| GROSS HYPOXIA VIA HIF1A DN
| 110 | 78 | All SZGR 2.0 genes in this pathway |
| GROSS HYPOXIA VIA ELK3 AND HIF1A UP
| 142 | 104 | All SZGR 2.0 genes in this pathway |
| SCHAEFFER PROSTATE DEVELOPMENT 6HR UP
| 176 | 115 | All SZGR 2.0 genes in this pathway |
| SHEN SMARCA2 TARGETS UP
| 424 | 268 | All SZGR 2.0 genes in this pathway |
| VANTVEER BREAST CANCER METASTASIS DN
| 121 | 65 | All SZGR 2.0 genes in this pathway |
| MANALO HYPOXIA UP
| 207 | 145 | All SZGR 2.0 genes in this pathway |
| FLECHNER BIOPSY KIDNEY TRANSPLANT REJECTED VS OK DN
| 546 | 351 | All SZGR 2.0 genes in this pathway |
| FLECHNER BIOPSY KIDNEY TRANSPLANT OK VS DONOR UP
| 555 | 346 | All SZGR 2.0 genes in this pathway |
| NADLER OBESITY DN
| 48 | 34 | All SZGR 2.0 genes in this pathway |
| MENSE HYPOXIA UP
| 98 | 71 | All SZGR 2.0 genes in this pathway |
| MARTINEZ RESPONSE TO TRABECTEDIN
| 50 | 32 | All SZGR 2.0 genes in this pathway |
| IVANOVA HEMATOPOIESIS LATE PROGENITOR
| 544 | 307 | All SZGR 2.0 genes in this pathway |
| BROWNE HCMV INFECTION 20HR DN
| 101 | 70 | All SZGR 2.0 genes in this pathway |
| KAAB HEART ATRIUM VS VENTRICLE DN
| 261 | 183 | All SZGR 2.0 genes in this pathway |
| GENTILE UV LOW DOSE DN
| 67 | 46 | All SZGR 2.0 genes in this pathway |
| BURTON ADIPOGENESIS 6
| 189 | 112 | All SZGR 2.0 genes in this pathway |
| ACEVEDO LIVER TUMOR VS NORMAL ADJACENT TISSUE UP
| 863 | 514 | All SZGR 2.0 genes in this pathway |
| WINTER HYPOXIA METAGENE
| 242 | 168 | All SZGR 2.0 genes in this pathway |
| RUIZ TNC TARGETS UP
| 153 | 107 | All SZGR 2.0 genes in this pathway |
| BOYAULT LIVER CANCER SUBCLASS G1 DN
| 40 | 26 | All SZGR 2.0 genes in this pathway |
| MOOTHA GLYCOGEN METABOLISM
| 21 | 16 | All SZGR 2.0 genes in this pathway |
| MARTENS BOUND BY PML RARA FUSION
| 456 | 287 | All SZGR 2.0 genes in this pathway |
| QI HYPOXIA
| 140 | 96 | All SZGR 2.0 genes in this pathway |
| JOHNSTONE PARVB TARGETS 2 DN
| 336 | 211 | All SZGR 2.0 genes in this pathway |
| PURBEY TARGETS OF CTBP1 NOT SATB1 UP
| 344 | 215 | All SZGR 2.0 genes in this pathway |
| KRIEG HYPOXIA NOT VIA KDM3A
| 770 | 480 | All SZGR 2.0 genes in this pathway |
| FORTSCHEGGER PHF8 TARGETS DN
| 784 | 464 | All SZGR 2.0 genes in this pathway |
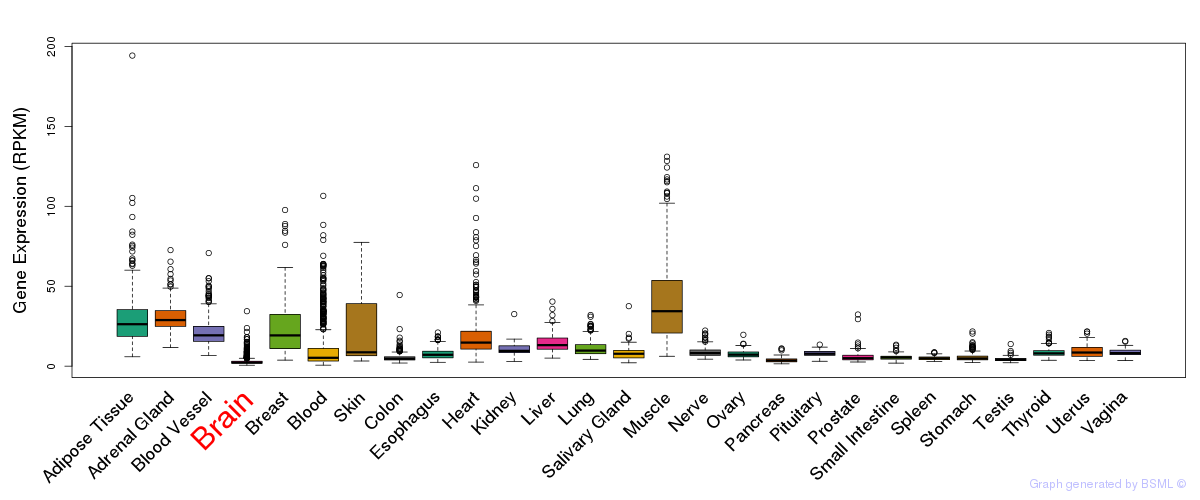
 Differentially methylated gene
Differentially methylated gene eQTL annotation
eQTL annotation