Gene Page: GBX2
Summary ?
| GeneID | 2637 |
| Symbol | GBX2 |
| Synonyms | - |
| Description | gastrulation brain homeobox 2 |
| Reference | MIM:601135|HGNC:HGNC:4186|Ensembl:ENSG00000168505|HPRD:03087|Vega:OTTHUMG00000133294 |
| Gene type | protein-coding |
| Map location | 2q37.2 |
| Pascal p-value | 0.172 |
| Fetal beta | 0.198 |
| DMG | 1 (# studies) |
| eGene | Myers' cis & trans |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CV:PGCnp | Genome-wide Association Study | GWAS | |
| DMG:Jaffe_2016 | Genome-wide DNA methylation analysis | This dataset includes 2,104 probes/CpGs associated with SZ patients (n=108) compared to 136 controls at Bonferroni-adjusted P < 0.05. | 1 |
| GO_Annotation | Mapping neuro-related keywords to Gene Ontology annotations | Hits with neuro-related keywords: 6 |
Section I. Genetics and epigenetics annotation
 Differentially methylated gene
Differentially methylated gene
| Probe | Chromosome | Position | Nearest gene | P (dis) | Beta (dis) | FDR (dis) | Study |
|---|---|---|---|---|---|---|---|
| cg18065680 | 2 | 237087147 | GBX2 | 1.51E-9 | -0.019 | 1.41E-6 | DMG:Jaffe_2016 |
 eQTL annotation
eQTL annotation
| SNP ID | Chromosome | Position | eGene | Gene Entrez ID | pvalue | qvalue | TSS distance | eQTL type |
|---|---|---|---|---|---|---|---|---|
| rs10929202 | chr2 | 237843949 | GBX2 | 2637 | 0.19 | cis |
Section II. Transcriptome annotation
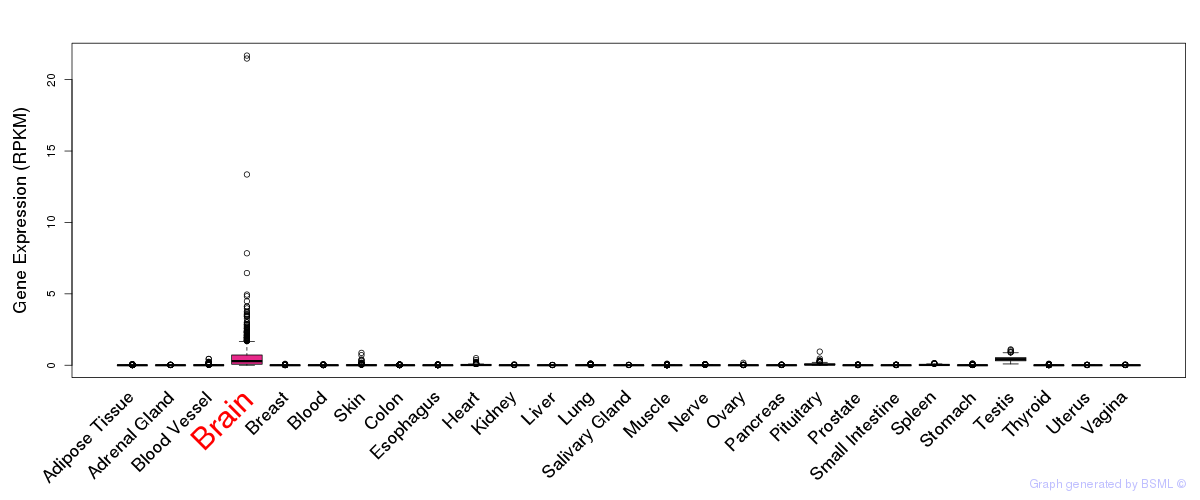
General gene expression (GTEx)
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
Top co-expressed genes in brain regions
| Top 10 positively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| SLC2A5 | 0.55 | 0.50 |
| MRPL41 | 0.55 | 0.53 |
| PTPMT1 | 0.54 | 0.59 |
| BRP44L | 0.54 | 0.56 |
| MSRB2 | 0.53 | 0.57 |
| DCXR | 0.53 | 0.55 |
| MTHFS | 0.53 | 0.56 |
| IL17D | 0.52 | 0.52 |
| C17orf89 | 0.51 | 0.49 |
| MID1IP1 | 0.51 | 0.42 |
| Top 10 negatively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| ZNF611 | -0.37 | -0.32 |
| AFF2 | -0.37 | -0.29 |
| ZNF91 | -0.36 | -0.32 |
| AC010522.1 | -0.36 | -0.38 |
| ZNF445 | -0.36 | -0.29 |
| MAP1B | -0.35 | -0.28 |
| RP11-468B6.3 | -0.35 | -0.25 |
| WNK3 | -0.35 | -0.24 |
| ZNF749 | -0.35 | -0.30 |
| AC011477.2 | -0.35 | -0.27 |
Section III. Gene Ontology annotation
| Molecular function | GO term | Evidence | Neuro keywords | PubMed ID |
|---|---|---|---|---|
| GO:0003700 | transcription factor activity | TAS | 9346236 | |
| GO:0043565 | sequence-specific DNA binding | IEA | - | |
| Biological process | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0007411 | axon guidance | IEA | axon (GO term level: 13) | - |
| GO:0030902 | hindbrain development | IEA | Brain (GO term level: 8) | - |
| GO:0021555 | midbrain-hindbrain boundary morphogenesis | IEA | Brain (GO term level: 11) | - |
| GO:0021549 | cerebellum development | IEA | Brain (GO term level: 10) | - |
| GO:0021930 | granule cell precursor proliferation | IEA | neuron, glutamate (GO term level: 11) | - |
| GO:0048483 | autonomic nervous system development | IEA | neuron (GO term level: 6) | - |
| GO:0001755 | neural crest cell migration | IEA | - | |
| GO:0001569 | patterning of blood vessels | IEA | - | |
| GO:0006355 | regulation of transcription, DNA-dependent | IEA | - | |
| GO:0008283 | cell proliferation | IEA | - | |
| GO:0042472 | inner ear morphogenesis | IEA | - | |
| GO:0021568 | rhombomere 2 development | IEA | - | |
| GO:0035239 | tube morphogenesis | IEA | - | |
| Cellular component | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0005634 | nucleus | IEA | - |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| TANG SENESCENCE TP53 TARGETS UP | 33 | 20 | All SZGR 2.0 genes in this pathway |
| NUYTTEN NIPP1 TARGETS DN | 848 | 527 | All SZGR 2.0 genes in this pathway |
| NUYTTEN EZH2 TARGETS DN | 1024 | 594 | All SZGR 2.0 genes in this pathway |
| BENPORATH SUZ12 TARGETS | 1038 | 678 | All SZGR 2.0 genes in this pathway |
| BENPORATH EED TARGETS | 1062 | 725 | All SZGR 2.0 genes in this pathway |
| BENPORATH ES WITH H3K27ME3 | 1118 | 744 | All SZGR 2.0 genes in this pathway |
| BENPORATH PRC2 TARGETS | 652 | 441 | All SZGR 2.0 genes in this pathway |
| TESAR ALK TARGETS HUMAN ES 5D UP | 5 | 5 | All SZGR 2.0 genes in this pathway |
| XU GH1 EXOGENOUS TARGETS DN | 120 | 69 | All SZGR 2.0 genes in this pathway |
| TESAR ALK TARGETS EPISC 3D UP | 7 | 7 | All SZGR 2.0 genes in this pathway |
| MEISSNER BRAIN HCP WITH H3K27ME3 | 269 | 159 | All SZGR 2.0 genes in this pathway |
| FIGUEROA AML METHYLATION CLUSTER 4 DN | 15 | 9 | All SZGR 2.0 genes in this pathway |
| FIGUEROA AML METHYLATION CLUSTER 6 DN | 38 | 19 | All SZGR 2.0 genes in this pathway |
| CHYLA CBFA2T3 TARGETS UP | 387 | 225 | All SZGR 2.0 genes in this pathway |
| HUANG GATA2 TARGETS DN | 72 | 52 | All SZGR 2.0 genes in this pathway |
| DELACROIX RAR BOUND ES | 462 | 273 | All SZGR 2.0 genes in this pathway |
| ZWANG TRANSIENTLY UP BY 2ND EGF PULSE ONLY | 1725 | 838 | All SZGR 2.0 genes in this pathway |
Section VI. microRNA annotation
| miRNA family | Target position | miRNA ID | miRNA seq | ||
|---|---|---|---|---|---|
| UTR start | UTR end | Match method | |||
| miR-369-3p | 207 | 213 | 1A | hsa-miR-369-3p | AAUAAUACAUGGUUGAUCUUU |
| miR-374 | 207 | 213 | m8 | hsa-miR-374 | UUAUAAUACAACCUGAUAAGUG |
- SZ: miRNAs which differentially expressed in brain cortex of schizophrenia patients comparing with control samples using microarray. Click here to see the list of SZ related miRNAs.
- Brain: miRNAs which are expressed in brain based on miRNA microarray expression studies. Click here to see the list of brain related miRNAs.