Gene Page: LAT
Summary ?
| GeneID | 27040 |
| Symbol | LAT |
| Synonyms | LAT1|pp36 |
| Description | linker for activation of T-cells |
| Reference | MIM:602354|HGNC:HGNC:18874|Ensembl:ENSG00000213658|HPRD:03832|Vega:OTTHUMG00000131761 |
| Gene type | protein-coding |
| Map location | 16p11.2 |
| Pascal p-value | 0.274 |
| Fetal beta | 0.013 |
| eGene | Myers' cis & trans |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CV:PGCnp | Genome-wide Association Study | GWAS | |
| PMID:cooccur | High-throughput literature-search | Systematic search in PubMed for genes co-occurring with SCZ keywords. A total of 3027 genes were included. | |
| GSMA_IIE | Genome scan meta-analysis (European-ancestry samples) | Psr: 0.01775 | |
| Literature | High-throughput literature-search | Co-occurance with Schizophrenia keywords: schizophrenia,schizophrenic,schizophrenics,schizophrenias,schizotypal | Click to show details |
| GO_Annotation | Mapping neuro-related keywords to Gene Ontology annotations | Hits with neuro-related keywords: 2 |
Section I. Genetics and epigenetics annotation
 eQTL annotation
eQTL annotation
| SNP ID | Chromosome | Position | eGene | Gene Entrez ID | pvalue | qvalue | TSS distance | eQTL type |
|---|---|---|---|---|---|---|---|---|
| rs6687849 | chr1 | 175904808 | LAT | 27040 | 0.15 | trans | ||
| rs2502827 | chr1 | 176044216 | LAT | 27040 | 0.01 | trans | ||
| rs16829545 | chr2 | 151977407 | LAT | 27040 | 0 | trans | ||
| rs3845734 | chr2 | 171125572 | LAT | 27040 | 0 | trans | ||
| rs7584986 | chr2 | 184111432 | LAT | 27040 | 2.313E-4 | trans | ||
| rs2090117 | chr4 | 54300590 | LAT | 27040 | 0.08 | trans | ||
| rs504836 | chr4 | 56285304 | LAT | 27040 | 0.13 | trans | ||
| rs534654 | chr4 | 56290219 | LAT | 27040 | 0.16 | trans | ||
| rs10491487 | chr5 | 80323367 | LAT | 27040 | 0.07 | trans | ||
| rs1368303 | chr5 | 147672388 | LAT | 27040 | 0.01 | trans | ||
| rs17263352 | chr9 | 124811572 | LAT | 27040 | 0.04 | trans | ||
| rs17104720 | chr14 | 77127308 | LAT | 27040 | 0.01 | trans | ||
| rs16955618 | chr15 | 29937543 | LAT | 27040 | 2.165E-9 | trans | ||
| rs749043 | chr20 | 46995700 | LAT | 27040 | 0.18 | trans |
Section II. Transcriptome annotation
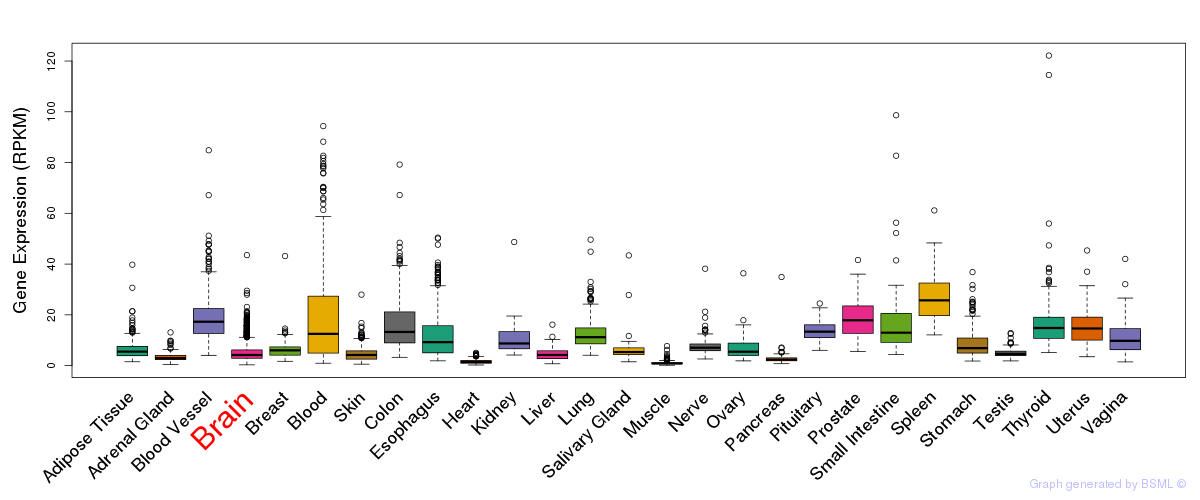
General gene expression (GTEx)
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
Top co-expressed genes in brain regions
| Top 10 positively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| CCDC8 | 0.82 | 0.49 |
| EFNA4 | 0.80 | 0.62 |
| NECAP2 | 0.80 | 0.62 |
| PDPN | 0.78 | 0.52 |
| RASSF10 | 0.77 | 0.52 |
| CD99 | 0.77 | 0.66 |
| ABHD4 | 0.77 | 0.64 |
| RFX2 | 0.76 | 0.51 |
| SIX5 | 0.76 | 0.62 |
| SULT1C4 | 0.75 | 0.62 |
| Top 10 negatively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| NDRG1 | -0.37 | -0.50 |
| FBXW7 | -0.37 | -0.50 |
| AC016910.1 | -0.36 | -0.53 |
| AC079953.2 | -0.34 | -0.50 |
| MEF2C | -0.34 | -0.51 |
| ARHGAP20 | -0.33 | -0.48 |
| DOCK9 | -0.32 | -0.44 |
| C1orf115 | -0.32 | -0.40 |
| LMO7 | -0.32 | -0.50 |
| DYNC1I1 | -0.32 | -0.48 |
Section III. Gene Ontology annotation
| Molecular function | GO term | Evidence | Neuro keywords | PubMed ID |
|---|---|---|---|---|
| GO:0005070 | SH3/SH2 adaptor activity | IDA | 9489702 | |
| GO:0005515 | protein binding | IPI | 16938345 | |
| Biological process | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0043303 | mast cell degranulation | IEA | serotonin (GO term level: 9) | - |
| GO:0007265 | Ras protein signal transduction | IMP | 12646565 | |
| GO:0007242 | intracellular signaling cascade | IDA | 12646565 | |
| GO:0007229 | integrin-mediated signaling pathway | IDA | 15100278 | |
| GO:0006955 | immune response | IDA | 15100278 | |
| GO:0019722 | calcium-mediated signaling | IMP | 12646565 | |
| GO:0050863 | regulation of T cell activation | IMP | 12646565 | |
| Cellular component | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0001772 | immunological synapse | IDA | Synap (GO term level: 7) | 12646565 |
| GO:0016021 | integral to membrane | IDA | 12646565 | |
| GO:0005886 | plasma membrane | EXP | 11048639 |11607830 |17652306 | |
| GO:0005886 | plasma membrane | IEA | - | |
| GO:0045121 | membrane raft | TAS | 14722116 |
Section IV. Protein-protein interaction annotation
| Interactors | Aliases B | Official full name B | Experimental | Source | PubMed ID |
|---|---|---|---|---|---|
| CD244 | 2B4 | NAIL | NKR2B4 | Nmrk | SLAMF4 | CD244 molecule, natural killer cell receptor 2B4 | - | HPRD | 12077228 |
| CD4 | CD4mut | CD4 molecule | - | HPRD | 10562325 |
| CD8A | CD8 | Leu2 | MAL | p32 | CD8a molecule | - | HPRD | 10562325 |
| CLYBL | CLB | bA134O15.1 | citrate lyase beta like | Affinity Capture-Western | BioGRID | 11368773 |
| FCGR1A | CD64 | CD64A | FCRI | FLJ18345 | IGFR1 | Fc fragment of IgG, high affinity Ia, receptor (CD64) | - | HPRD,BioGRID | 10781611 |
| FCGR2A | CD32 | CD32A | CDw32 | FCG2 | FCGR2 | FCGR2A1 | FcGR | IGFR2 | MGC23887 | MGC30032 | Fc fragment of IgG, low affinity IIa, receptor (CD32) | - | HPRD,BioGRID | 10781611 |
| GAB2 | KIAA0571 | GRB2-associated binding protein 2 | - | HPRD,BioGRID | 11572860 |
| GRAP | MGC64880 | GRB2-related adaptor protein | Affinity Capture-Western Reconstituted Complex | BioGRID | 9489702 |12186560 |
| GRAP2 | GADS | GRAP-2 | GRB2L | GRBLG | GRID | GRPL | GrbX | Grf40 | Mona | P38 | GRB2-related adaptor protein 2 | - | HPRD,BioGRID | 10021361 |
| GRB2 | ASH | EGFRBP-GRB2 | Grb3-3 | MST084 | MSTP084 | growth factor receptor-bound protein 2 | - | HPRD | 9489702 |12359715 |
| GRB2 | ASH | EGFRBP-GRB2 | Grb3-3 | MST084 | MSTP084 | growth factor receptor-bound protein 2 | Affinity Capture-Western Reconstituted Complex | BioGRID | 9489702 |11368773 |12186560 |
| ITK | EMT | LYK | MGC126257 | MGC126258 | PSCTK2 | IL2-inducible T-cell kinase | - | HPRD | 10506192|12186560 |
| ITK | EMT | LYK | MGC126257 | MGC126258 | PSCTK2 | IL2-inducible T-cell kinase | Affinity Capture-Western Biochemical Activity | BioGRID | 10506192 |12186560 |
| LCP2 | SLP-76 | SLP76 | lymphocyte cytosolic protein 2 (SH2 domain containing leukocyte protein of 76kDa) | - | HPRD | 9489702 |
| PIK3R1 | GRB1 | p85 | p85-ALPHA | phosphoinositide-3-kinase, regulatory subunit 1 (alpha) | Affinity Capture-Western Reconstituted Complex | BioGRID | 11368773 |
| PLCG1 | PLC-II | PLC1 | PLC148 | PLCgamma1 | phospholipase C, gamma 1 | - | HPRD,BioGRID | 9729044 |11368773 |
| PLCG1 | PLC-II | PLC1 | PLC148 | PLCgamma1 | phospholipase C, gamma 1 | LAT interacts with an unspecified isoform of PLC-gamma-1. | BIND | 15696170 |
| PLCG2 | - | phospholipase C, gamma 2 (phosphatidylinositol-specific) | - | HPRD,BioGRID | 10469124 |
| PTPRJ | CD148 | DEP1 | HPTPeta | R-PTP-ETA | SCC1 | protein tyrosine phosphatase, receptor type, J | - | HPRD,BioGRID | 11259588 |
| SH3BP2 | 3BP2 | CRBM | CRPM | FLJ42079 | RES4-23 | SH3-domain binding protein 2 | - | HPRD,BioGRID | 11390470 |
| SHB | RP11-3J10.8 | bA3J10.2 | Src homology 2 domain containing adaptor protein B | - | HPRD,BioGRID | 12084069 |
| SLA | SLA1 | SLAP | Src-like-adaptor | - | HPRD,BioGRID | 10449770 |
| SYK | DKFZp313N1010 | FLJ25043 | FLJ37489 | spleen tyrosine kinase | Biochemical Activity | BioGRID | 11368773 |
| SYK | DKFZp313N1010 | FLJ25043 | FLJ37489 | spleen tyrosine kinase | - | HPRD | 9891970 |
| VAV1 | VAV | vav 1 guanine nucleotide exchange factor | - | HPRD,BioGRID | 11368773 |
| ZAP70 | FLJ17670 | FLJ17679 | SRK | STD | TZK | ZAP-70 | zeta-chain (TCR) associated protein kinase 70kDa | Biochemical Activity | BioGRID | 11368773 |12186560 |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| KEGG NATURAL KILLER CELL MEDIATED CYTOTOXICITY | 137 | 92 | All SZGR 2.0 genes in this pathway |
| KEGG T CELL RECEPTOR SIGNALING PATHWAY | 108 | 89 | All SZGR 2.0 genes in this pathway |
| KEGG FC EPSILON RI SIGNALING PATHWAY | 79 | 58 | All SZGR 2.0 genes in this pathway |
| KEGG FC GAMMA R MEDIATED PHAGOCYTOSIS | 97 | 71 | All SZGR 2.0 genes in this pathway |
| BIOCARTA NKCELLS PATHWAY | 20 | 13 | All SZGR 2.0 genes in this pathway |
| BIOCARTA TCR PATHWAY | 49 | 37 | All SZGR 2.0 genes in this pathway |
| ST T CELL SIGNAL TRANSDUCTION | 45 | 33 | All SZGR 2.0 genes in this pathway |
| PID FCER1 PATHWAY | 62 | 43 | All SZGR 2.0 genes in this pathway |
| PID TCR PATHWAY | 66 | 51 | All SZGR 2.0 genes in this pathway |
| PID PTP1B PATHWAY | 52 | 40 | All SZGR 2.0 genes in this pathway |
| PID CD8 TCR PATHWAY | 53 | 42 | All SZGR 2.0 genes in this pathway |
| PID PI3KCI PATHWAY | 49 | 40 | All SZGR 2.0 genes in this pathway |
| PID TCR JNK PATHWAY | 14 | 11 | All SZGR 2.0 genes in this pathway |
| REACTOME TCR SIGNALING | 54 | 46 | All SZGR 2.0 genes in this pathway |
| REACTOME GENERATION OF SECOND MESSENGER MOLECULES | 27 | 25 | All SZGR 2.0 genes in this pathway |
| REACTOME GPVI MEDIATED ACTIVATION CASCADE | 31 | 25 | All SZGR 2.0 genes in this pathway |
| REACTOME HEMOSTASIS | 466 | 331 | All SZGR 2.0 genes in this pathway |
| REACTOME IMMUNE SYSTEM | 933 | 616 | All SZGR 2.0 genes in this pathway |
| REACTOME ADAPTIVE IMMUNE SYSTEM | 539 | 350 | All SZGR 2.0 genes in this pathway |
| REACTOME PLATELET ACTIVATION SIGNALING AND AGGREGATION | 208 | 138 | All SZGR 2.0 genes in this pathway |
| KIM MYC AMPLIFICATION TARGETS UP | 201 | 127 | All SZGR 2.0 genes in this pathway |
| NUYTTEN EZH2 TARGETS DN | 1024 | 594 | All SZGR 2.0 genes in this pathway |
| MOREIRA RESPONSE TO TSA DN | 18 | 16 | All SZGR 2.0 genes in this pathway |
| QI PLASMACYTOMA UP | 259 | 185 | All SZGR 2.0 genes in this pathway |
| VILIMAS NOTCH1 TARGETS UP | 52 | 41 | All SZGR 2.0 genes in this pathway |
| MIKKELSEN ES ICP WITH H3K4ME3 | 718 | 401 | All SZGR 2.0 genes in this pathway |
| HUANG GATA2 TARGETS DN | 72 | 52 | All SZGR 2.0 genes in this pathway |
| KRIEG HYPOXIA NOT VIA KDM3A | 770 | 480 | All SZGR 2.0 genes in this pathway |
| BOSCO INTERFERON INDUCED ANTIVIRAL MODULE | 78 | 48 | All SZGR 2.0 genes in this pathway |