Gene Page: GOLIM4
Summary ?
| GeneID | 27333 |
| Symbol | GOLIM4 |
| Synonyms | GIMPC|GOLPH4|GPP130|P138 |
| Description | golgi integral membrane protein 4 |
| Reference | MIM:606805|HGNC:HGNC:15448|Ensembl:ENSG00000173905|HPRD:08429|Vega:OTTHUMG00000158554 |
| Gene type | protein-coding |
| Map location | 3q26.2 |
| Pascal p-value | 0.017 |
| Sherlock p-value | 0.386 |
| Fetal beta | 0.959 |
| DMG | 1 (# studies) |
| eGene | Myers' cis & trans |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CV:PGCnp | Genome-wide Association Study | GWAS | |
| DMG:Jaffe_2016 | Genome-wide DNA methylation analysis | This dataset includes 2,104 probes/CpGs associated with SZ patients (n=108) compared to 136 controls at Bonferroni-adjusted P < 0.05. | 1 |
Section I. Genetics and epigenetics annotation
 Differentially methylated gene
Differentially methylated gene
| Probe | Chromosome | Position | Nearest gene | P (dis) | Beta (dis) | FDR (dis) | Study |
|---|---|---|---|---|---|---|---|
| cg20620272 | 3 | 167968129 | GOLIM4 | 2.68E-8 | -0.014 | 8.51E-6 | DMG:Jaffe_2016 |
 eQTL annotation
eQTL annotation
| SNP ID | Chromosome | Position | eGene | Gene Entrez ID | pvalue | qvalue | TSS distance | eQTL type |
|---|---|---|---|---|---|---|---|---|
| rs1707266 | chr1 | 211554194 | GOLIM4 | 27333 | 0.12 | trans | ||
| rs994871 | chr2 | 188999579 | GOLIM4 | 27333 | 0.16 | trans | ||
| rs6737451 | chr2 | 189039508 | GOLIM4 | 27333 | 0.02 | trans | ||
| rs1395163 | chr3 | 31028951 | GOLIM4 | 27333 | 4.938E-5 | trans | ||
| rs17029291 | chr3 | 32402138 | GOLIM4 | 27333 | 0 | trans | ||
| rs624496 | chr3 | 120506736 | GOLIM4 | 27333 | 0.01 | trans | ||
| rs17030258 | chr4 | 154546102 | GOLIM4 | 27333 | 0.05 | trans | ||
| rs16870339 | chr5 | 2705169 | GOLIM4 | 27333 | 0.02 | trans | ||
| rs4976298 | chr5 | 134780729 | GOLIM4 | 27333 | 0.19 | trans | ||
| rs4617108 | chr7 | 49423831 | GOLIM4 | 27333 | 0.03 | trans | ||
| snp_a-1904613 | 0 | GOLIM4 | 27333 | 0.01 | trans | |||
| rs4624992 | chr8 | 11291596 | GOLIM4 | 27333 | 0.17 | trans | ||
| rs7039404 | chr9 | 90934986 | GOLIM4 | 27333 | 0.11 | trans | ||
| rs11259374 | chr10 | 14917473 | GOLIM4 | 27333 | 0.03 | trans | ||
| rs11009196 | chr10 | 33334853 | GOLIM4 | 27333 | 0.04 | trans | ||
| rs909288 | chr10 | 124257417 | GOLIM4 | 27333 | 0 | trans | ||
| rs11021829 | chr11 | 11416216 | GOLIM4 | 27333 | 0.19 | trans | ||
| rs2031706 | chr12 | 100899659 | GOLIM4 | 27333 | 0.04 | trans | ||
| rs2254834 | chr14 | 43596184 | GOLIM4 | 27333 | 0.12 | trans | ||
| rs1954396 | chr14 | 43601835 | GOLIM4 | 27333 | 0.01 | trans | ||
| rs4641819 | chr18 | 45911492 | GOLIM4 | 27333 | 0.09 | trans | ||
| rs5999295 | chr22 | 34726495 | GOLIM4 | 27333 | 0.09 | trans | ||
| rs5999296 | chr22 | 34727119 | GOLIM4 | 27333 | 0.08 | trans | ||
| rs12157904 | chr22 | 37982011 | GOLIM4 | 27333 | 0.02 | trans | ||
| rs986774 | chrX | 7573775 | GOLIM4 | 27333 | 0.08 | trans |
Section II. Transcriptome annotation
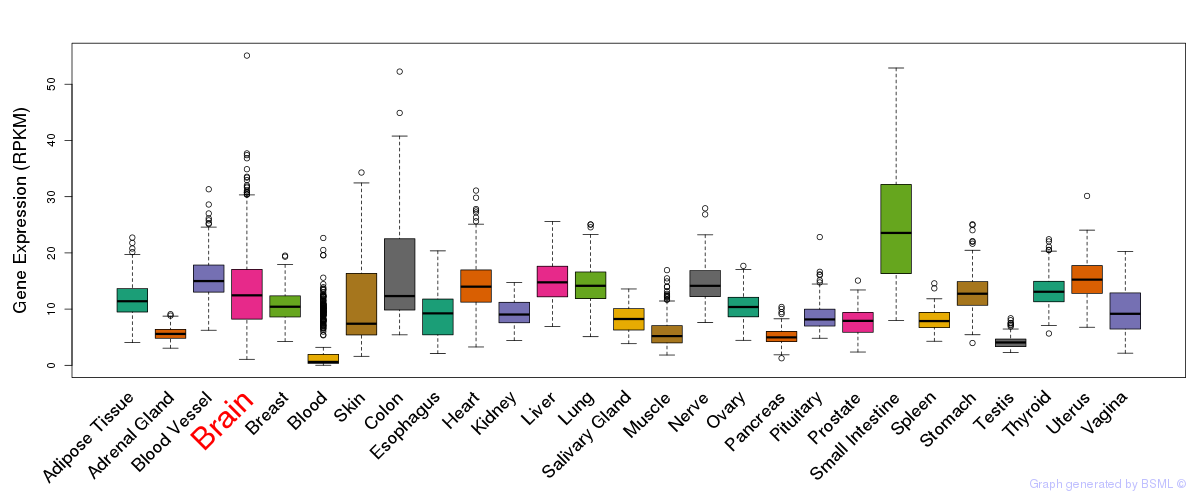
General gene expression (GTEx)
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
No co-expressed genes in brain regions
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| OSMAN BLADDER CANCER UP | 404 | 246 | All SZGR 2.0 genes in this pathway |
| ZHOU INFLAMMATORY RESPONSE FIMA UP | 544 | 308 | All SZGR 2.0 genes in this pathway |
| KIM WT1 TARGETS 8HR UP | 164 | 122 | All SZGR 2.0 genes in this pathway |
| JAATINEN HEMATOPOIETIC STEM CELL UP | 316 | 190 | All SZGR 2.0 genes in this pathway |
| GRAESSMANN APOPTOSIS BY DOXORUBICIN DN | 1781 | 1082 | All SZGR 2.0 genes in this pathway |
| HAMAI APOPTOSIS VIA TRAIL UP | 584 | 356 | All SZGR 2.0 genes in this pathway |
| BEGUM TARGETS OF PAX3 FOXO1 FUSION UP | 60 | 45 | All SZGR 2.0 genes in this pathway |
| GRUETZMANN PANCREATIC CANCER UP | 358 | 245 | All SZGR 2.0 genes in this pathway |
| PUJANA CHEK2 PCC NETWORK | 779 | 480 | All SZGR 2.0 genes in this pathway |
| NUYTTEN EZH2 TARGETS DN | 1024 | 594 | All SZGR 2.0 genes in this pathway |
| SCHAEFFER PROSTATE DEVELOPMENT 6HR DN | 514 | 330 | All SZGR 2.0 genes in this pathway |
| BENPORATH NANOG TARGETS | 988 | 594 | All SZGR 2.0 genes in this pathway |
| GOLDRATH HOMEOSTATIC PROLIFERATION | 171 | 102 | All SZGR 2.0 genes in this pathway |
| GOLDRATH IMMUNE MEMORY | 65 | 42 | All SZGR 2.0 genes in this pathway |
| KUMAR TARGETS OF MLL AF9 FUSION | 405 | 264 | All SZGR 2.0 genes in this pathway |
| IVANOVA HEMATOPOIESIS MATURE CELL | 293 | 160 | All SZGR 2.0 genes in this pathway |
| BLALOCK ALZHEIMERS DISEASE UP | 1691 | 1088 | All SZGR 2.0 genes in this pathway |
| BROWNE HCMV INFECTION 48HR DN | 504 | 323 | All SZGR 2.0 genes in this pathway |
| CREIGHTON ENDOCRINE THERAPY RESISTANCE 1 | 528 | 324 | All SZGR 2.0 genes in this pathway |
| IZADPANAH STEM CELL ADIPOSE VS BONE UP | 126 | 92 | All SZGR 2.0 genes in this pathway |
| JOHNSTONE PARVB TARGETS 3 DN | 918 | 550 | All SZGR 2.0 genes in this pathway |
| JUBAN TARGETS OF SPI1 AND FLI1 UP | 115 | 73 | All SZGR 2.0 genes in this pathway |
| LIM MAMMARY STEM CELL UP | 489 | 314 | All SZGR 2.0 genes in this pathway |