Gene Page: GMDS
Summary ?
| GeneID | 2762 |
| Symbol | GMDS |
| Synonyms | GMD|SDR3E1 |
| Description | GDP-mannose 4,6-dehydratase |
| Reference | MIM:602884|HGNC:HGNC:4369|Ensembl:ENSG00000112699|HPRD:11925|Vega:OTTHUMG00000016143 |
| Gene type | protein-coding |
| Map location | 6p25 |
| Pascal p-value | 0.361 |
| Sherlock p-value | 0.852 |
| Fetal beta | -0.303 |
| DMG | 3 (# studies) |
| eGene | Myers' cis & trans |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CV:PGCnp | Genome-wide Association Study | GWAS | |
| DMG:Jaffe_2016 | Genome-wide DNA methylation analysis | This dataset includes 2,104 probes/CpGs associated with SZ patients (n=108) compared to 136 controls at Bonferroni-adjusted P < 0.05. | 3 |
| DMG:Montano_2016 | Genome-wide DNA methylation analysis | This dataset includes 172 replicated associations between CpGs with schizophrenia. | 3 |
| DMG:Wockner_2014 | Genome-wide DNA methylation analysis | This dataset includes 4641 differentially methylated probes corresponding to 2929 unique genes between schizophrenia patients (n=24) and controls (n=24). | 3 |
| GSMA_I | Genome scan meta-analysis | Psr: 0.0159 |
Section I. Genetics and epigenetics annotation
 Differentially methylated gene
Differentially methylated gene
| Probe | Chromosome | Position | Nearest gene | P (dis) | Beta (dis) | FDR (dis) | Study |
|---|---|---|---|---|---|---|---|
| cg06315217 | 6 | 1629850 | GMDS | 4.51E-5 | 0.006 | 0.093 | DMG:Montano_2016 |
| cg11786839 | 6 | 1635578 | GMDS | 3.28E-6 | 0.474 | 0.009 | DMG:Wockner_2014 |
| cg08932320 | 6 | 2246077 | GMDS | 1.07E-8 | -0.015 | 4.57E-6 | DMG:Jaffe_2016 |
 eQTL annotation
eQTL annotation
| SNP ID | Chromosome | Position | eGene | Gene Entrez ID | pvalue | qvalue | TSS distance | eQTL type |
|---|---|---|---|---|---|---|---|---|
| rs17135501 | chr6 | 2704474 | GMDS | 2762 | 0.15 | cis | ||
| rs17135521 | chr6 | 2707631 | GMDS | 2762 | 0.15 | cis | ||
| rs11706314 | chr3 | 45309716 | GMDS | 2762 | 0.11 | trans | ||
| rs2922180 | chr3 | 125280472 | GMDS | 2762 | 0.01 | trans | ||
| rs2971271 | chr3 | 125450402 | GMDS | 2762 | 0.12 | trans | ||
| rs2976809 | chr3 | 125451738 | GMDS | 2762 | 1.767E-4 | trans | ||
| rs7668550 | chr4 | 104384355 | GMDS | 2762 | 0 | trans | ||
| rs6449550 | chr5 | 61034312 | GMDS | 2762 | 0.13 | trans | ||
| rs9313797 | chr5 | 158465457 | GMDS | 2762 | 0.07 | trans | ||
| rs17056474 | chr5 | 158468105 | GMDS | 2762 | 0.07 | trans | ||
| rs9321275 | chr6 | 131493217 | GMDS | 2762 | 0.04 | trans | ||
| rs2199402 | chr8 | 9201002 | GMDS | 2762 | 0.01 | trans | ||
| rs7841407 | chr8 | 9243427 | GMDS | 2762 | 0 | trans | ||
| rs2061799 | chr9 | 97795003 | GMDS | 2762 | 0.08 | trans | ||
| rs11067174 | chr12 | 115011927 | GMDS | 2762 | 0.06 | trans | ||
| rs9529974 | chr13 | 72821935 | GMDS | 2762 | 0.14 | trans | ||
| rs17720221 | chr13 | 75182377 | GMDS | 2762 | 0.01 | trans | ||
| rs7165622 | chr15 | 93979910 | GMDS | 2762 | 0.05 | trans | ||
| rs11873703 | chr18 | 61448702 | GMDS | 2762 | 0.01 | trans | ||
| rs6095741 | chr20 | 48666589 | GMDS | 2762 | 0.01 | trans |
Section II. Transcriptome annotation
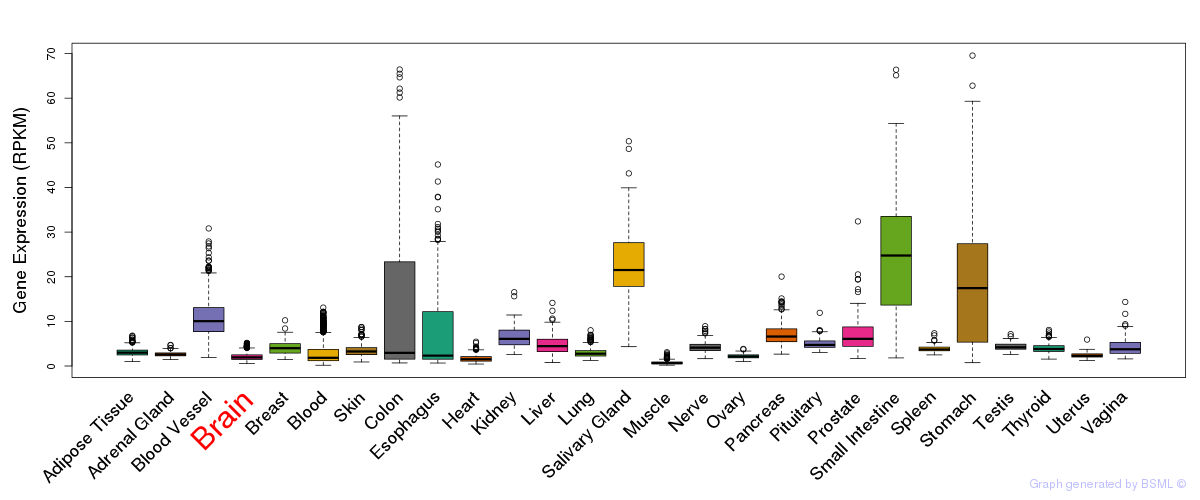
General gene expression (GTEx)
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
Top co-expressed genes in brain regions
| Top 10 positively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| TTC16 | 0.34 | 0.23 |
| GUCY2D | 0.33 | 0.23 |
| IL28RA | 0.33 | 0.11 |
| EYA2 | 0.32 | 0.26 |
| ATBF1 | 0.31 | 0.10 |
| KCNIP1 | 0.31 | 0.17 |
| TSHZ2 | 0.31 | 0.14 |
| TNFRSF11B | 0.31 | 0.29 |
| ALK | 0.30 | 0.14 |
| C13orf29 | 0.30 | 0.19 |
| Top 10 negatively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| GPR22 | -0.25 | -0.29 |
| EMX1 | -0.23 | -0.25 |
| GTDC1 | -0.22 | -0.20 |
| C6orf115 | -0.21 | -0.24 |
| CIDEA | -0.21 | -0.26 |
| B3GALT2 | -0.21 | -0.20 |
| AC021078.3 | -0.21 | -0.27 |
| NEUROD6 | -0.21 | -0.24 |
| IER5L | -0.21 | -0.20 |
| SEH1L | -0.20 | -0.22 |
Section III. Gene Ontology annotation
| Molecular function | GO term | Evidence | Neuro keywords | PubMed ID |
|---|---|---|---|---|
| GO:0003824 | catalytic activity | IEA | - | |
| GO:0005488 | binding | IEA | - | |
| GO:0016829 | lyase activity | IEA | - | |
| GO:0008446 | GDP-mannose 4,6-dehydratase activity | IEA | - | |
| GO:0008446 | GDP-mannose 4,6-dehydratase activity | TAS | 11698403 | |
| GO:0016491 | oxidoreductase activity | IEA | - | |
| GO:0050662 | coenzyme binding | IEA | - | |
| Biological process | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0005975 | carbohydrate metabolic process | TAS | 9525924 | |
| GO:0008152 | metabolic process | IEA | - | |
| GO:0007159 | leukocyte adhesion | NAS | 11698403 | |
| GO:0042351 | 'de novo' GDP-L-fucose biosynthetic process | TAS | 11698403 | |
| GO:0019673 | GDP-mannose metabolic process | IEA | - | |
| GO:0044237 | cellular metabolic process | IEA | - | |
| Cellular component | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0005622 | intracellular | IEA | - | |
| GO:0005737 | cytoplasm | IC | 11698403 |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| KEGG FRUCTOSE AND MANNOSE METABOLISM | 34 | 23 | All SZGR 2.0 genes in this pathway |
| KEGG AMINO SUGAR AND NUCLEOTIDE SUGAR METABOLISM | 44 | 30 | All SZGR 2.0 genes in this pathway |
| NAKAMURA TUMOR ZONE PERIPHERAL VS CENTRAL DN | 634 | 384 | All SZGR 2.0 genes in this pathway |
| LIU PROSTATE CANCER UP | 96 | 57 | All SZGR 2.0 genes in this pathway |
| ONKEN UVEAL MELANOMA DN | 526 | 357 | All SZGR 2.0 genes in this pathway |
| CHEMNITZ RESPONSE TO PROSTAGLANDIN E2 DN | 391 | 222 | All SZGR 2.0 genes in this pathway |
| DIAZ CHRONIC MEYLOGENOUS LEUKEMIA UP | 1382 | 904 | All SZGR 2.0 genes in this pathway |
| DOANE RESPONSE TO ANDROGEN UP | 184 | 125 | All SZGR 2.0 genes in this pathway |
| VECCHI GASTRIC CANCER ADVANCED VS EARLY DN | 138 | 70 | All SZGR 2.0 genes in this pathway |
| WAMUNYOKOLI OVARIAN CANCER LMP UP | 265 | 158 | All SZGR 2.0 genes in this pathway |
| WAMUNYOKOLI OVARIAN CANCER GRADES 1 2 UP | 137 | 84 | All SZGR 2.0 genes in this pathway |
| DODD NASOPHARYNGEAL CARCINOMA UP | 1821 | 933 | All SZGR 2.0 genes in this pathway |
| GAUSSMANN MLL AF4 FUSION TARGETS A UP | 191 | 128 | All SZGR 2.0 genes in this pathway |
| KOYAMA SEMA3B TARGETS UP | 292 | 168 | All SZGR 2.0 genes in this pathway |
| BENPORATH MYC MAX TARGETS | 775 | 494 | All SZGR 2.0 genes in this pathway |
| AMIT EGF RESPONSE 480 HELA | 164 | 118 | All SZGR 2.0 genes in this pathway |
| PENG RAPAMYCIN RESPONSE UP | 203 | 130 | All SZGR 2.0 genes in this pathway |
| IVANOVA HEMATOPOIESIS EARLY PROGENITOR | 532 | 309 | All SZGR 2.0 genes in this pathway |
| REN ALVEOLAR RHABDOMYOSARCOMA DN | 408 | 274 | All SZGR 2.0 genes in this pathway |
| CUI TCF21 TARGETS 2 DN | 830 | 547 | All SZGR 2.0 genes in this pathway |
| GAJATE RESPONSE TO TRABECTEDIN DN | 19 | 11 | All SZGR 2.0 genes in this pathway |
| BAELDE DIABETIC NEPHROPATHY DN | 434 | 302 | All SZGR 2.0 genes in this pathway |
| KRIGE RESPONSE TO TOSEDOSTAT 6HR UP | 953 | 554 | All SZGR 2.0 genes in this pathway |
| KRIGE RESPONSE TO TOSEDOSTAT 24HR DN | 1011 | 592 | All SZGR 2.0 genes in this pathway |
| CREIGHTON ENDOCRINE THERAPY RESISTANCE 3 | 720 | 440 | All SZGR 2.0 genes in this pathway |
| MARSON BOUND BY FOXP3 UNSTIMULATED | 1229 | 713 | All SZGR 2.0 genes in this pathway |
| ACEVEDO LIVER TUMOR VS NORMAL ADJACENT TISSUE UP | 863 | 514 | All SZGR 2.0 genes in this pathway |
| SMID BREAST CANCER BASAL UP | 648 | 398 | All SZGR 2.0 genes in this pathway |
| IZADPANAH STEM CELL ADIPOSE VS BONE UP | 126 | 92 | All SZGR 2.0 genes in this pathway |
| GRADE COLON CANCER UP | 871 | 505 | All SZGR 2.0 genes in this pathway |
| CROMER TUMORIGENESIS DN | 51 | 29 | All SZGR 2.0 genes in this pathway |
| YAGI AML WITH 11Q23 REARRANGED | 351 | 238 | All SZGR 2.0 genes in this pathway |
| DANG BOUND BY MYC | 1103 | 714 | All SZGR 2.0 genes in this pathway |
| CHICAS RB1 TARGETS CONFLUENT | 567 | 365 | All SZGR 2.0 genes in this pathway |
| JOHNSTONE PARVB TARGETS 2 DN | 336 | 211 | All SZGR 2.0 genes in this pathway |
| ACEVEDO FGFR1 TARGETS IN PROSTATE CANCER MODEL UP | 289 | 184 | All SZGR 2.0 genes in this pathway |