Gene Page: GNB2
Summary ?
| GeneID | 2783 |
| Symbol | GNB2 |
| Synonyms | - |
| Description | G protein subunit beta 2 |
| Reference | MIM:139390|HGNC:HGNC:4398|Ensembl:ENSG00000172354|HPRD:11820|Vega:OTTHUMG00000137419 |
| Gene type | protein-coding |
| Map location | 7q22 |
| Pascal p-value | 4.787E-4 |
| Sherlock p-value | 0.078 |
| TADA p-value | 0.011 |
| Fetal beta | 0.294 |
| DMG | 4 (# studies) |
| eGene | Myers' cis & trans Meta |
| Support | CANABINOID DOPAMINE G-PROTEIN RELAY METABOTROPIC GLUTAMATE RECEPTOR SEROTONIN G2Cdb.human_BAYES-COLLINS-HUMAN-PSD-CONSENSUS G2Cdb.human_BAYES-COLLINS-HUMAN-PSD-FULL G2Cdb.human_BAYES-COLLINS-MOUSE-PSD-CONSENSUS G2Cdb.human_clathrin G2Cdb.human_Synaptosome G2Cdb.humanNRC G2Cdb.humanPSD G2Cdb.humanPSP CompositeSet |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CV:PGCnp | Genome-wide Association Study | GWAS | |
| DMG:Jaffe_2016 | Genome-wide DNA methylation analysis | This dataset includes 2,104 probes/CpGs associated with SZ patients (n=108) compared to 136 controls at Bonferroni-adjusted P < 0.05. | 5 |
| DMG:Nishioka_2013 | Genome-wide DNA methylation analysis | The authors investigated the methylation profiles of DNA in peripheral blood cells from 18 patients with first-episode schizophrenia (FESZ) and from 15 normal controls. | 5 |
| DMG:vanEijk_2014 | Genome-wide DNA methylation analysis | This dataset includes 432 differentially methylated CpG sites corresponding to 391 unique transcripts between schizophrenia patients (n=260) and unaffected controls (n=250). | 5 |
| DMG:Wockner_2014 | Genome-wide DNA methylation analysis | This dataset includes 4641 differentially methylated probes corresponding to 2929 unique genes between schizophrenia patients (n=24) and controls (n=24). | 5 |
| DNM:Fromer_2014 | Whole Exome Sequencing analysis | This study reported a WES study of 623 schizophrenia trios, reporting DNMs using genomic DNA. |
Section I. Genetics and epigenetics annotation
 DNM table
DNM table
| Gene | Chromosome | Position | Ref | Alt | Transcript | AA change | Mutation type | Sift | CG46 | Trait | Study |
|---|---|---|---|---|---|---|---|---|---|---|---|
| GNB2 | chr7 | 100276061 | A | C | NM_005273 | p.247D>A | missense | Schizophrenia | DNM:Fromer_2014 |
 Differentially methylated gene
Differentially methylated gene
| Probe | Chromosome | Position | Nearest gene | P (dis) | Beta (dis) | FDR (dis) | Study |
|---|---|---|---|---|---|---|---|
| cg01399255 | 7 | 100271274 | GNB2 | 1.602E-4 | -0.244 | 0.032 | DMG:Wockner_2014 |
| cg11223864 | 7 | 100271323 | GNB2 | -0.04 | 0.3 | DMG:Nishioka_2013 | |
| cg11223864 | 7 | 100271323 | GNB2 | 1.85E-8 | -0.01 | 6.6E-6 | DMG:Jaffe_2016 |
| cg13650156 | 7 | 99970502 | GNB2 | 1.49E-5 | -4.307 | DMG:vanEijk_2014 | |
| cg01274660 | 7 | 100465625 | GNB2 | 6.497E-4 | -4.343 | DMG:vanEijk_2014 |
 eQTL annotation
eQTL annotation
| SNP ID | Chromosome | Position | eGene | Gene Entrez ID | pvalue | qvalue | TSS distance | eQTL type |
|---|---|---|---|---|---|---|---|---|
| rs9993882 | chr4 | 183824780 | GNB2 | 2783 | 0.11 | trans | ||
| rs10016530 | chr4 | 183824983 | GNB2 | 2783 | 0.11 | trans |
Section II. Transcriptome annotation
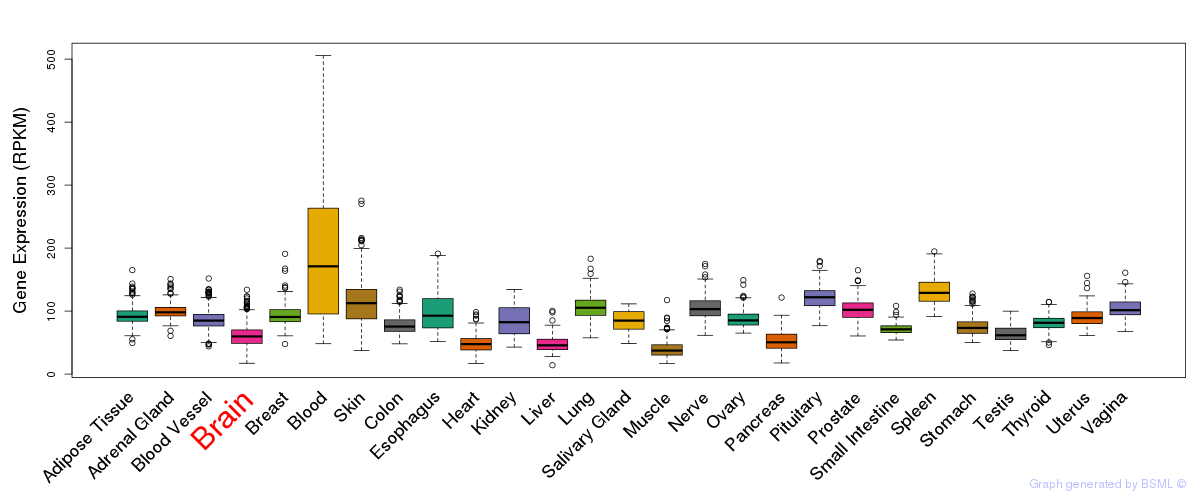
General gene expression (GTEx)
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
Top co-expressed genes in brain regions
| Top 10 positively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| NSUN2 | 0.89 | 0.90 |
| ZC3H7A | 0.88 | 0.87 |
| ZC3H11A | 0.87 | 0.90 |
| INTS3 | 0.87 | 0.88 |
| CLIP1 | 0.87 | 0.88 |
| INO80 | 0.87 | 0.89 |
| USP48 | 0.86 | 0.88 |
| CCDC93 | 0.86 | 0.90 |
| BRWD2 | 0.86 | 0.87 |
| VPS54 | 0.86 | 0.89 |
| Top 10 negatively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| AF347015.31 | -0.72 | -0.74 |
| AF347015.21 | -0.70 | -0.76 |
| MT-CO2 | -0.70 | -0.72 |
| AF347015.27 | -0.68 | -0.69 |
| IFI27 | -0.68 | -0.68 |
| HIGD1B | -0.67 | -0.70 |
| AF347015.8 | -0.66 | -0.69 |
| MT-CYB | -0.66 | -0.67 |
| AF347015.33 | -0.64 | -0.64 |
| FXYD1 | -0.63 | -0.64 |
Section III. Gene Ontology annotation
| Molecular function | GO term | Evidence | Neuro keywords | PubMed ID |
|---|---|---|---|---|
| GO:0004871 | signal transducer activity | IEA | - | |
| GO:0003924 | GTPase activity | TAS | 2902634 | |
| Biological process | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0007186 | G-protein coupled receptor protein signaling pathway | TAS | 7649993 | |
| GO:0007165 | signal transduction | IEA | - | |
| GO:0009755 | hormone-mediated signaling | EXP | 12626323 |
Section IV. Protein-protein interaction annotation
| Interactors | Aliases B | Official full name B | Experimental | Source | PubMed ID |
|---|---|---|---|---|---|
| CAPNS1 | 30K | CALPAIN4 | CANP | CANPS | CAPN4 | CDPS | calpain, small subunit 1 | Two-hybrid | BioGRID | 16169070 |
| DNAJC25 | bA16L21.2.1 | DnaJ (Hsp40) homolog, subfamily C , member 25 | - | HPRD | 7665596 |
| FANCA | FA | FA-H | FA1 | FAA | FACA | FAH | FANCH | MGC75158 | Fanconi anemia, complementation group A | Two-hybrid | BioGRID | 14499622 |
| GNG10 | - | guanine nucleotide binding protein (G protein), gamma 10 | Reconstituted Complex | BioGRID | 7665596 |
| GNG12 | FLJ31352 | FLJ34695 | guanine nucleotide binding protein (G protein), gamma 12 | - | HPRD | 10409705 |
| GNG13 | G(gamma)13 | h2-35 | guanine nucleotide binding protein (G protein), gamma 13 | - | HPRD | 11675383 |
| GNG4 | DKFZp547K1018 | FLJ23803 | FLJ34187 | guanine nucleotide binding protein (G protein), gamma 4 | - | HPRD | 7665596 |
| GNG7 | FLJ00058 | guanine nucleotide binding protein (G protein), gamma 7 | - | HPRD,BioGRID | 8308009 |
| MAP3K3 | MAPKKK3 | MEKK3 | mitogen-activated protein kinase kinase kinase 3 | - | HPRD | 14743216 |
| PCBP1 | HNRPE1 | HNRPX | hnRNP-E1 | hnRNP-X | poly(rC) binding protein 1 | - | HPRD | 11591653 |
| RAF1 | CRAF | NS5 | Raf-1 | c-Raf | v-raf-1 murine leukemia viral oncogene homolog 1 | Affinity Capture-Western | BioGRID | 7782277 |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| KEGG CHEMOKINE SIGNALING PATHWAY | 190 | 128 | All SZGR 2.0 genes in this pathway |
| REACTOME GASTRIN CREB SIGNALLING PATHWAY VIA PKC AND MAPK | 205 | 136 | All SZGR 2.0 genes in this pathway |
| REACTOME TRANSMISSION ACROSS CHEMICAL SYNAPSES | 186 | 155 | All SZGR 2.0 genes in this pathway |
| REACTOME NEURONAL SYSTEM | 279 | 221 | All SZGR 2.0 genes in this pathway |
| REACTOME SIGNALING BY GPCR | 920 | 449 | All SZGR 2.0 genes in this pathway |
| REACTOME INTEGRATION OF ENERGY METABOLISM | 120 | 84 | All SZGR 2.0 genes in this pathway |
| REACTOME OPIOID SIGNALLING | 78 | 56 | All SZGR 2.0 genes in this pathway |
| REACTOME NEUROTRANSMITTER RECEPTOR BINDING AND DOWNSTREAM TRANSMISSION IN THE POSTSYNAPTIC CELL | 137 | 110 | All SZGR 2.0 genes in this pathway |
| REACTOME G PROTEIN ACTIVATION | 27 | 15 | All SZGR 2.0 genes in this pathway |
| REACTOME TRANSMEMBRANE TRANSPORT OF SMALL MOLECULES | 413 | 270 | All SZGR 2.0 genes in this pathway |
| REACTOME GLUCAGON SIGNALING IN METABOLIC REGULATION | 34 | 23 | All SZGR 2.0 genes in this pathway |
| REACTOME REGULATION OF INSULIN SECRETION BY GLUCAGON LIKE PEPTIDE1 | 43 | 28 | All SZGR 2.0 genes in this pathway |
| REACTOME G ALPHA Q SIGNALLING EVENTS | 184 | 116 | All SZGR 2.0 genes in this pathway |
| REACTOME REGULATION OF INSULIN SECRETION | 93 | 65 | All SZGR 2.0 genes in this pathway |
| REACTOME INHIBITION OF INSULIN SECRETION BY ADRENALINE NORADRENALINE | 25 | 15 | All SZGR 2.0 genes in this pathway |
| REACTOME CLASS B 2 SECRETIN FAMILY RECEPTORS | 88 | 58 | All SZGR 2.0 genes in this pathway |
| REACTOME GLUCAGON TYPE LIGAND RECEPTORS | 33 | 22 | All SZGR 2.0 genes in this pathway |
| REACTOME G ALPHA1213 SIGNALLING EVENTS | 74 | 56 | All SZGR 2.0 genes in this pathway |
| REACTOME ADP SIGNALLING THROUGH P2RY1 | 25 | 16 | All SZGR 2.0 genes in this pathway |
| REACTOME G BETA GAMMA SIGNALLING THROUGH PLC BETA | 20 | 12 | All SZGR 2.0 genes in this pathway |
| REACTOME GPCR DOWNSTREAM SIGNALING | 805 | 368 | All SZGR 2.0 genes in this pathway |
| REACTOME G ALPHA I SIGNALLING EVENTS | 195 | 114 | All SZGR 2.0 genes in this pathway |
| REACTOME G BETA GAMMA SIGNALLING THROUGH PI3KGAMMA | 25 | 15 | All SZGR 2.0 genes in this pathway |
| REACTOME G ALPHA S SIGNALLING EVENTS | 121 | 82 | All SZGR 2.0 genes in this pathway |
| REACTOME G ALPHA Z SIGNALLING EVENTS | 44 | 29 | All SZGR 2.0 genes in this pathway |
| REACTOME G PROTEIN BETA GAMMA SIGNALLING | 28 | 18 | All SZGR 2.0 genes in this pathway |
| REACTOME SIGNAL AMPLIFICATION | 31 | 19 | All SZGR 2.0 genes in this pathway |
| REACTOME THROMBOXANE SIGNALLING THROUGH TP RECEPTOR | 23 | 14 | All SZGR 2.0 genes in this pathway |
| REACTOME ADP SIGNALLING THROUGH P2RY12 | 21 | 10 | All SZGR 2.0 genes in this pathway |
| REACTOME ACTIVATION OF KAINATE RECEPTORS UPON GLUTAMATE BINDING | 31 | 23 | All SZGR 2.0 genes in this pathway |
| REACTOME GPCR LIGAND BINDING | 408 | 246 | All SZGR 2.0 genes in this pathway |
| REACTOME THROMBIN SIGNALLING THROUGH PROTEINASE ACTIVATED RECEPTORS PARS | 32 | 20 | All SZGR 2.0 genes in this pathway |
| REACTOME PLATELET HOMEOSTASIS | 78 | 49 | All SZGR 2.0 genes in this pathway |
| REACTOME AQUAPORIN MEDIATED TRANSPORT | 51 | 34 | All SZGR 2.0 genes in this pathway |
| REACTOME PROSTACYCLIN SIGNALLING THROUGH PROSTACYCLIN RECEPTOR | 19 | 11 | All SZGR 2.0 genes in this pathway |
| REACTOME REGULATION OF WATER BALANCE BY RENAL AQUAPORINS | 44 | 30 | All SZGR 2.0 genes in this pathway |
| REACTOME INHIBITION OF VOLTAGE GATED CA2 CHANNELS VIA GBETA GAMMA SUBUNITS | 25 | 17 | All SZGR 2.0 genes in this pathway |
| REACTOME GABA B RECEPTOR ACTIVATION | 38 | 26 | All SZGR 2.0 genes in this pathway |
| REACTOME GABA RECEPTOR ACTIVATION | 52 | 40 | All SZGR 2.0 genes in this pathway |
| REACTOME HEMOSTASIS | 466 | 331 | All SZGR 2.0 genes in this pathway |
| REACTOME POTASSIUM CHANNELS | 98 | 68 | All SZGR 2.0 genes in this pathway |
| REACTOME INWARDLY RECTIFYING K CHANNELS | 31 | 20 | All SZGR 2.0 genes in this pathway |
| REACTOME PLATELET ACTIVATION SIGNALING AND AGGREGATION | 208 | 138 | All SZGR 2.0 genes in this pathway |
| GARY CD5 TARGETS UP | 473 | 314 | All SZGR 2.0 genes in this pathway |
| DIAZ CHRONIC MEYLOGENOUS LEUKEMIA UP | 1382 | 904 | All SZGR 2.0 genes in this pathway |
| ENK UV RESPONSE KERATINOCYTE UP | 530 | 342 | All SZGR 2.0 genes in this pathway |
| SCHLOSSER SERUM RESPONSE UP | 134 | 93 | All SZGR 2.0 genes in this pathway |
| DAIRKEE TERT TARGETS UP | 380 | 213 | All SZGR 2.0 genes in this pathway |
| LOCKWOOD AMPLIFIED IN LUNG CANCER | 214 | 139 | All SZGR 2.0 genes in this pathway |
| MATSUDA NATURAL KILLER DIFFERENTIATION | 475 | 313 | All SZGR 2.0 genes in this pathway |
| THEILGAARD NEUTROPHIL AT SKIN WOUND DN | 225 | 163 | All SZGR 2.0 genes in this pathway |
| WEIGEL OXIDATIVE STRESS BY HNE AND H2O2 | 39 | 29 | All SZGR 2.0 genes in this pathway |
| LEE AGING CEREBELLUM UP | 84 | 58 | All SZGR 2.0 genes in this pathway |
| JAZAERI BREAST CANCER BRCA1 VS BRCA2 UP | 49 | 28 | All SZGR 2.0 genes in this pathway |
| ZAMORA NOS2 TARGETS DN | 96 | 71 | All SZGR 2.0 genes in this pathway |
| GRADE COLON CANCER UP | 871 | 505 | All SZGR 2.0 genes in this pathway |
| AGUIRRE PANCREATIC CANCER COPY NUMBER UP | 298 | 174 | All SZGR 2.0 genes in this pathway |
| MARTENS BOUND BY PML RARA FUSION | 456 | 287 | All SZGR 2.0 genes in this pathway |
| SMIRNOV RESPONSE TO IR 6HR UP | 166 | 97 | All SZGR 2.0 genes in this pathway |
Section VI. microRNA annotation
| miRNA family | Target position | miRNA ID | miRNA seq | ||
|---|---|---|---|---|---|
| UTR start | UTR end | Match method | |||
| miR-128 | 159 | 165 | m8 | hsa-miR-128a | UCACAGUGAACCGGUCUCUUUU |
| hsa-miR-128b | UCACAGUGAACCGGUCUCUUUC | ||||
| miR-142-3p | 50 | 57 | 1A,m8 | hsa-miR-142-3p | UGUAGUGUUUCCUACUUUAUGGA |
| miR-331 | 281 | 287 | m8 | hsa-miR-331brain | GCCCCUGGGCCUAUCCUAGAA |
| miR-369-3p | 322 | 328 | 1A | hsa-miR-369-3p | AAUAAUACAUGGUUGAUCUUU |
| miR-374 | 322 | 328 | m8 | hsa-miR-374 | UUAUAAUACAACCUGAUAAGUG |
| miR-490 | 133 | 139 | m8 | hsa-miR-490 | CAACCUGGAGGACUCCAUGCUG |
- SZ: miRNAs which differentially expressed in brain cortex of schizophrenia patients comparing with control samples using microarray. Click here to see the list of SZ related miRNAs.
- Brain: miRNAs which are expressed in brain based on miRNA microarray expression studies. Click here to see the list of brain related miRNAs.